2. Quick Tour of GraphSpace¶
2.1. Welcome Screen¶
The welcome page greets a user when the user visits GraphSpace. The Welcome Screen is designed to access commonly used features of GraphSpace like:
2.1.1. Create Account¶
The user can create an account on GraphSpace by following the given steps:
- Click on the
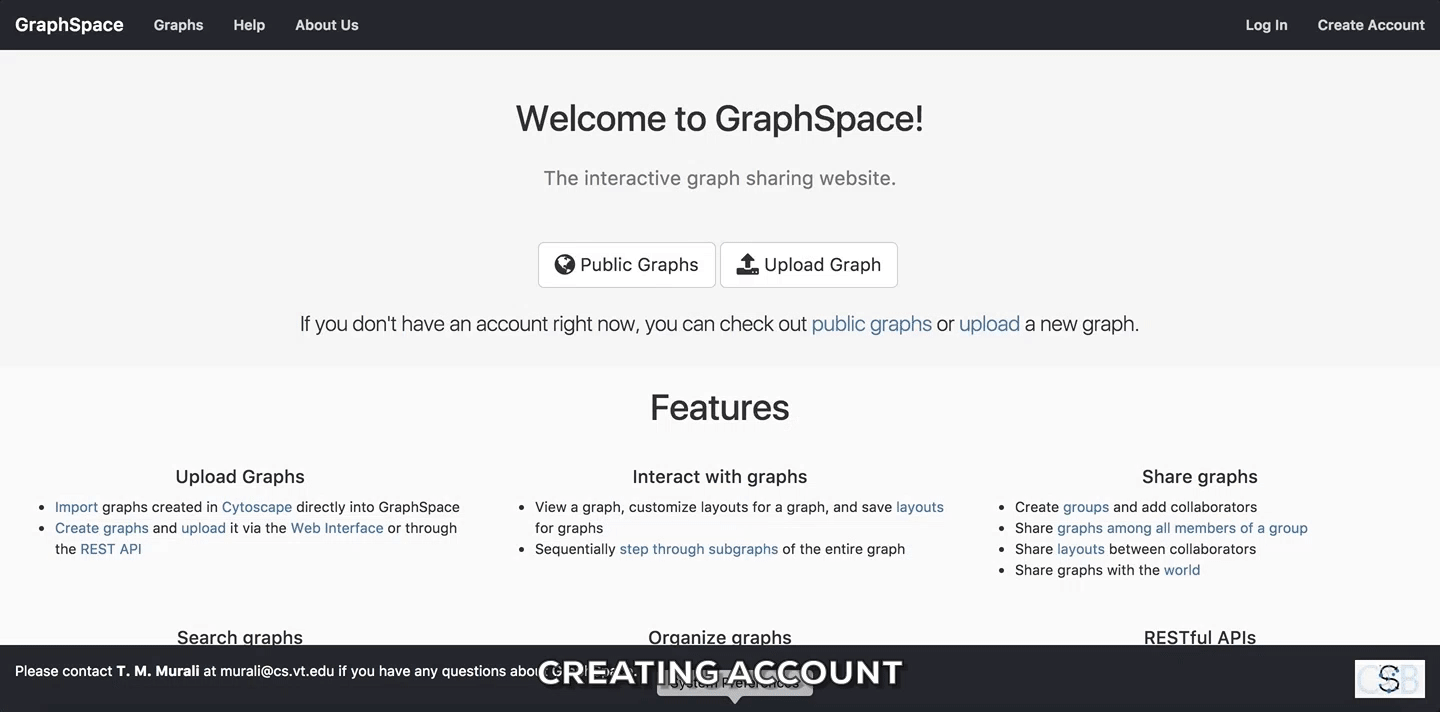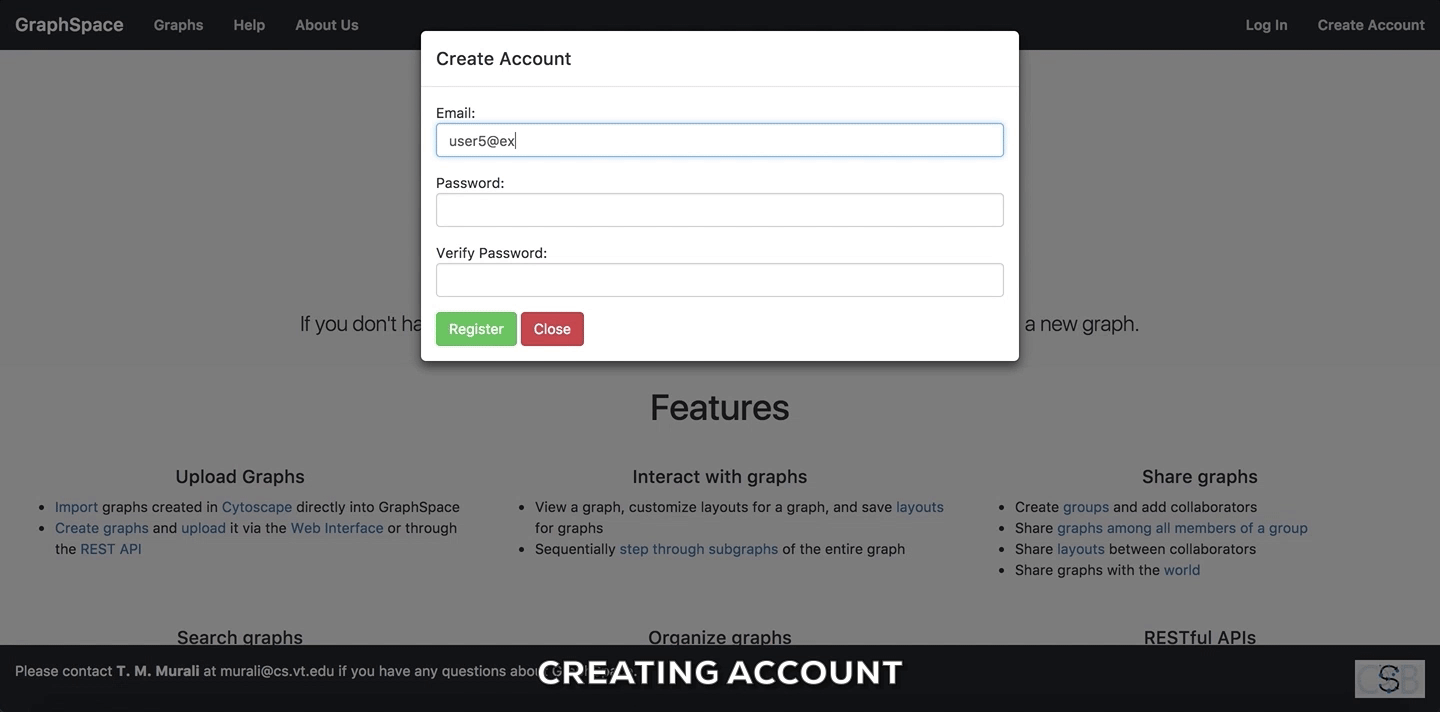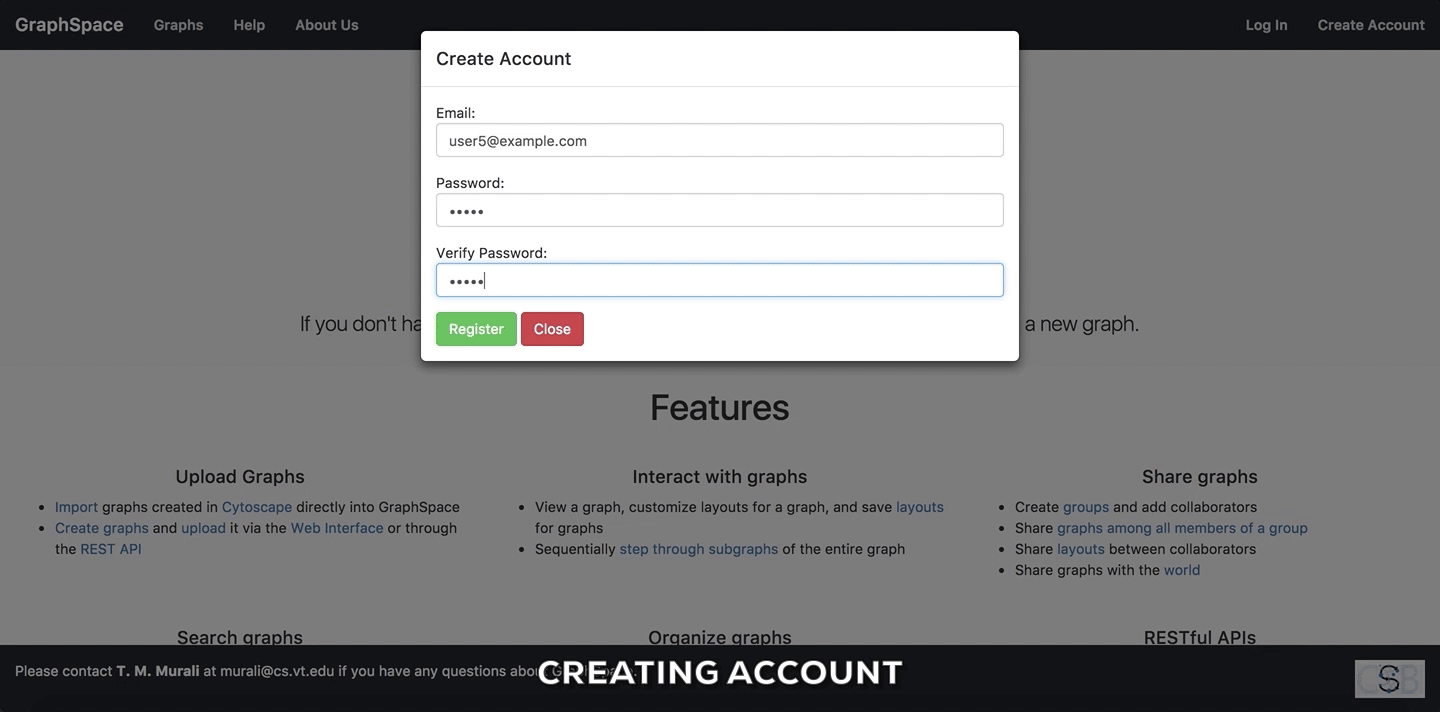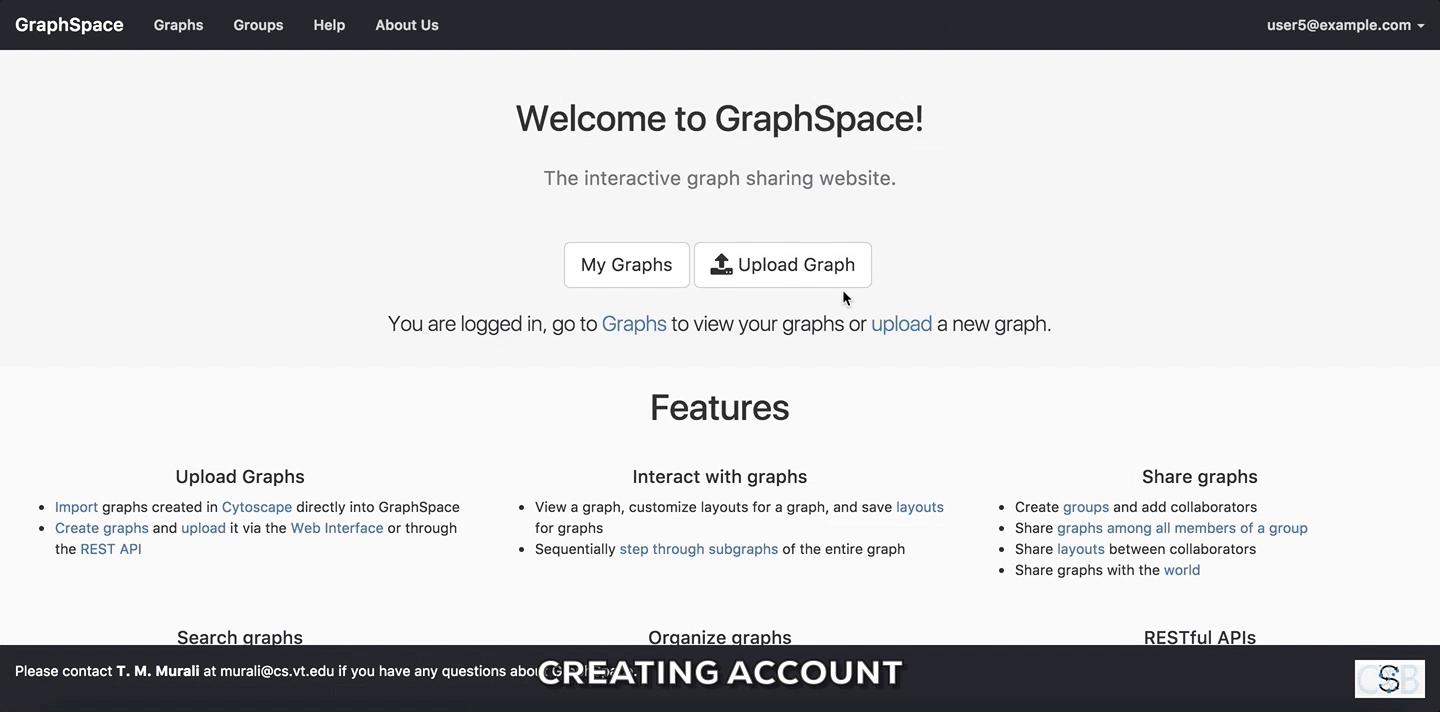
Create Accountbutton on the top navigation bar. This will trigger a create account window/pop-up to be displayed. - Enter the your email address.
- Enter a password for your account.
- Verify your password by entering the same password again.
- Click on
Submitbutton to create the account with the given email address and password.
 Create Account
Create Account
2.1.2. Log In¶
The user can log-in to GraphSpace by following the given steps:
- Click on the
Log Inbutton on the top navigation bar. This will trigger a log-in window/pop-up to be displayed. - Enter your email address.
- Enter you password.
- Click on
Submitbutton to log in with the given email address. - If you have forgotten you password, click on the
Forgot Passwordlink.
 Log In
Log In
For the rest of the tutorial, we assume that the user has already created an account. We also assume that the user belongs to a group with shared networks or that the user uploaded networks via the upload page or the RESTful API. The tutorial specifically steps through how user1@example.com interacts with GraphSpace after logging in.
2.1.3. Upload a graph¶
The user can upload a graph on GraphSpace by following the given steps:
- Go to the Upload Graph Page by clicking on the
Upload Graphbutton on Home Page. - Enter a unique name for the new graph.
- Select a CYJS file which contains the graph information.
- Select a JSON file which contains the style information for the graph. (Optional Step)
- Click on
Submitbutton to upload the graph using the selected files. - Once the graph has been uploaded, GraphSpace will provide a unique URL through which the user may interact with the graph represented by the uploaded files.
You can use the following sample files to learn how to upload graphs to GraphSpace:
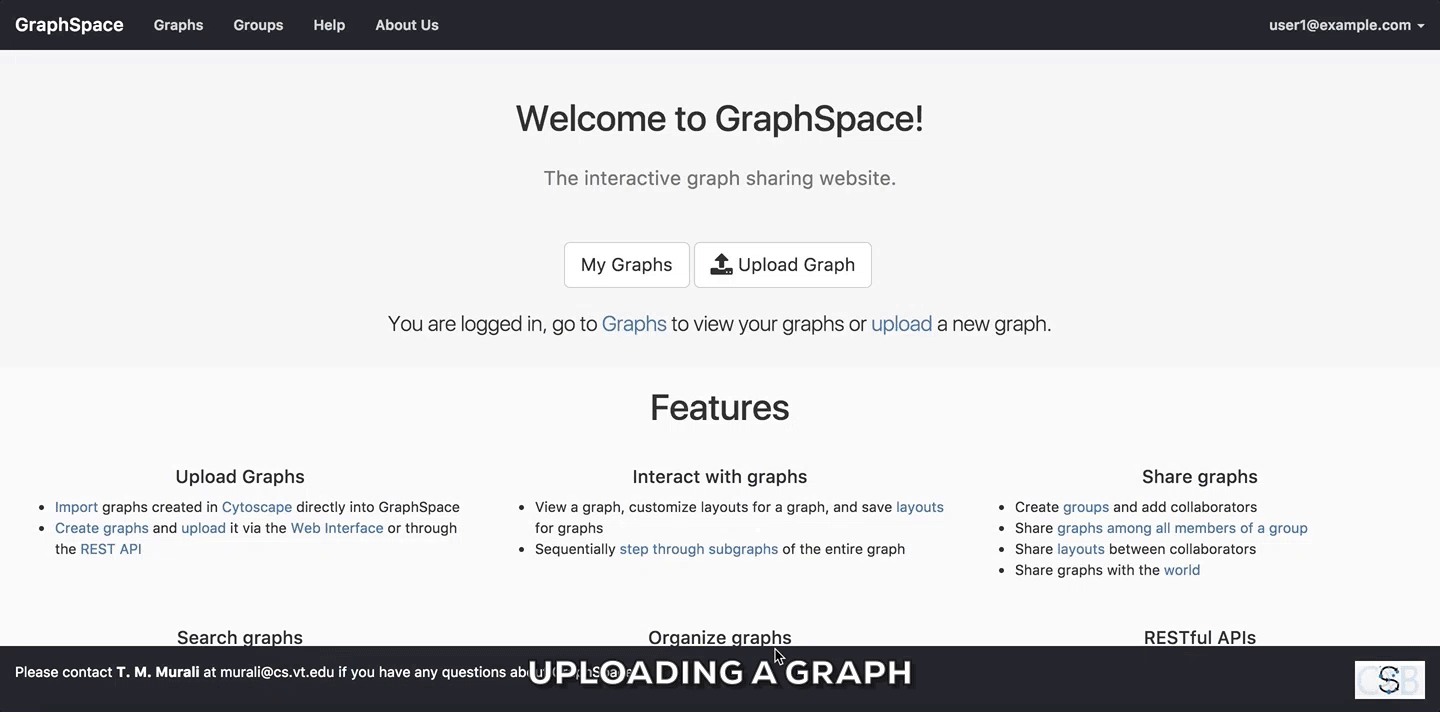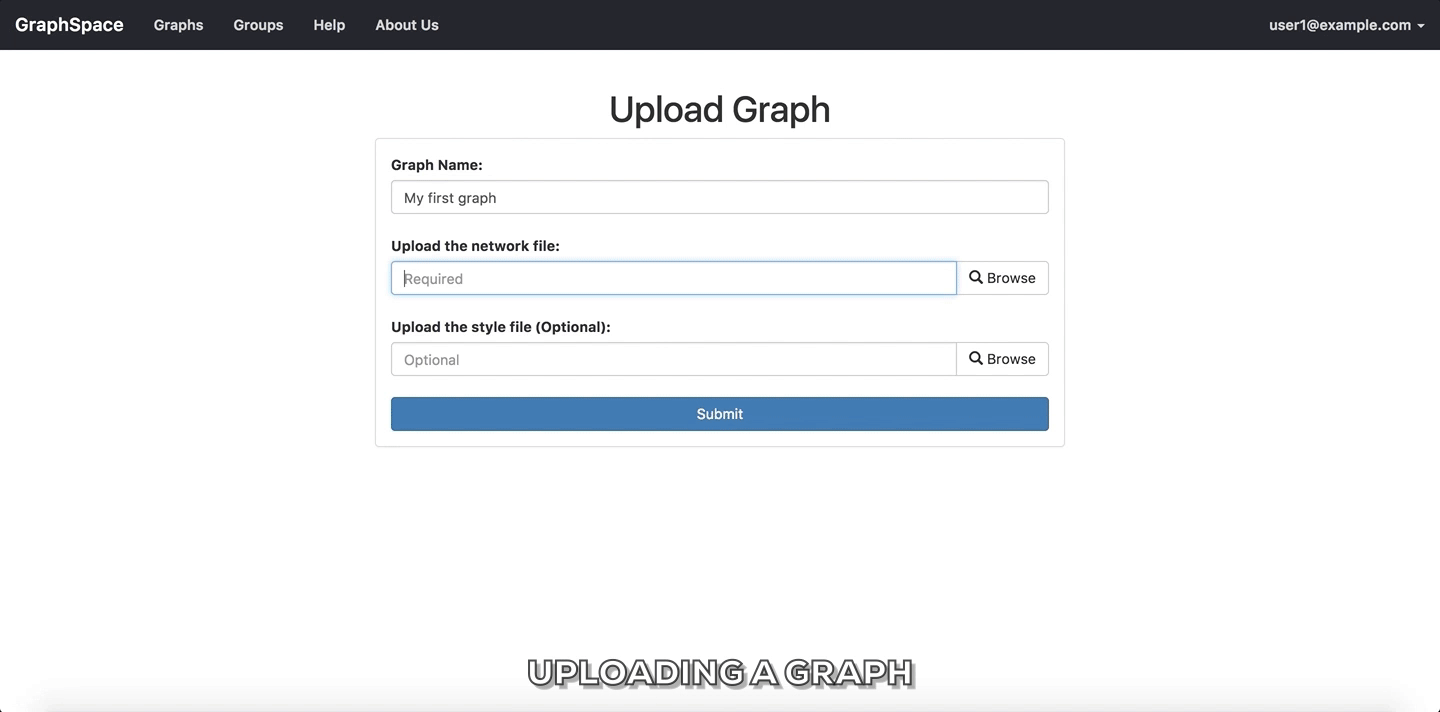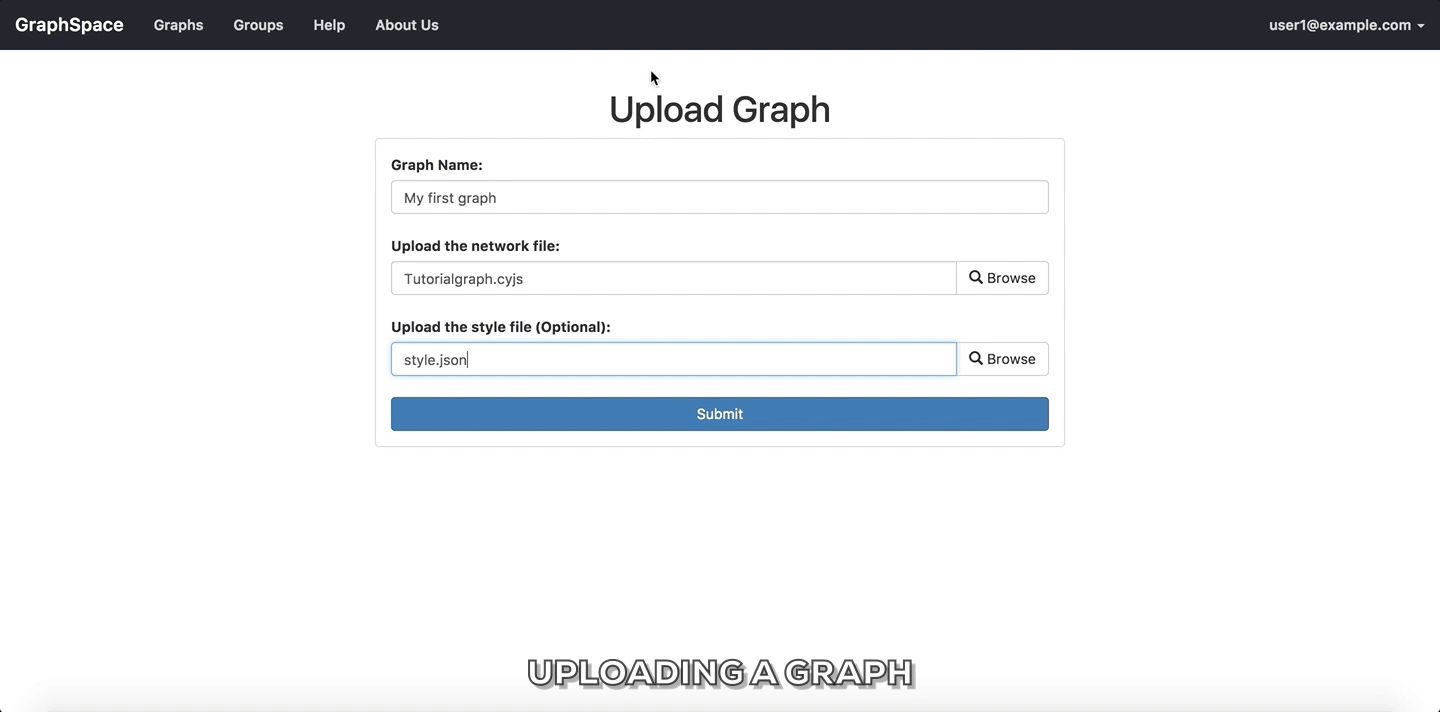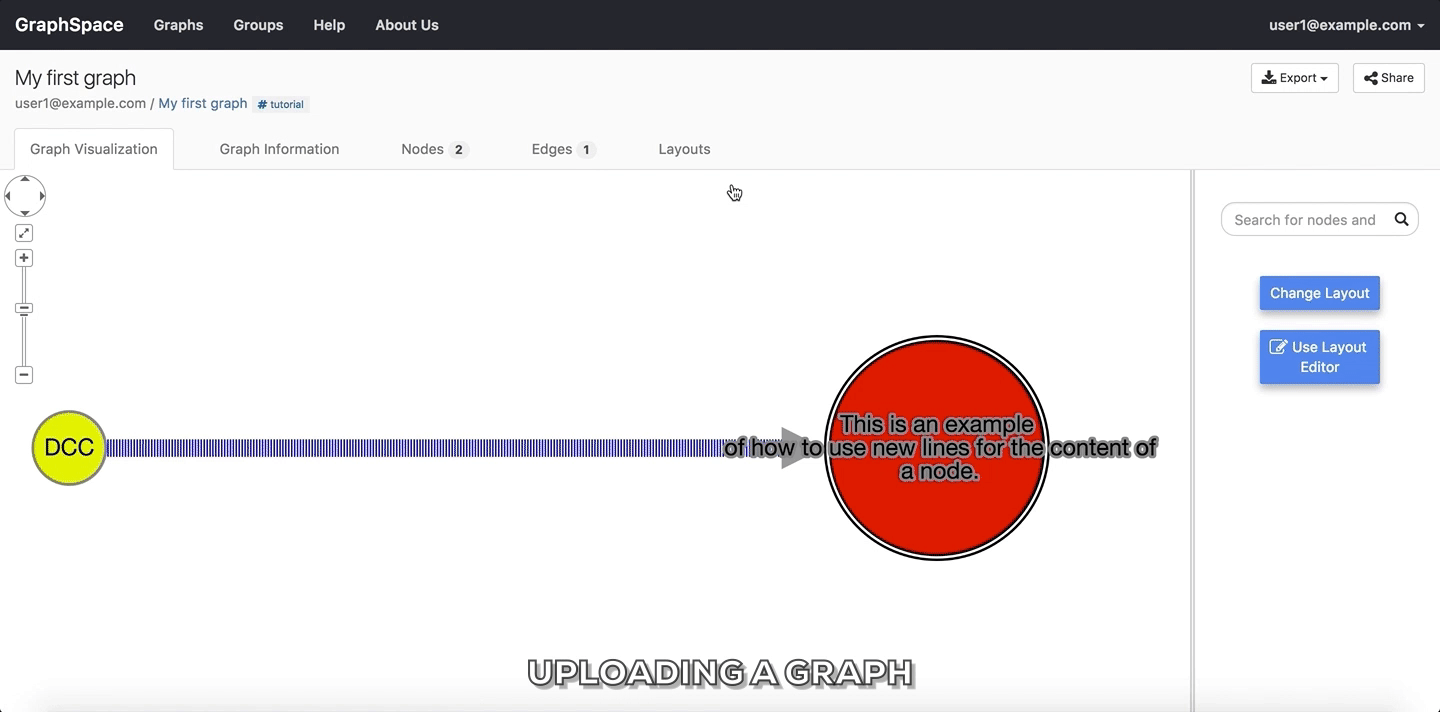 Upload a graph
Upload a graph
2.1.4. List of uploaded graphs¶
The user can go to a page that lists the graphs accessible by the user on GraphSpace by following the given steps:
- Click on the button titled
Graphson the top navigation bar. OR - Click on the
My Graphsbutton on the Home Page.
In this example, the user owns 33 graphs, can access 64 public graphs and 33 graphs are shared with this user.
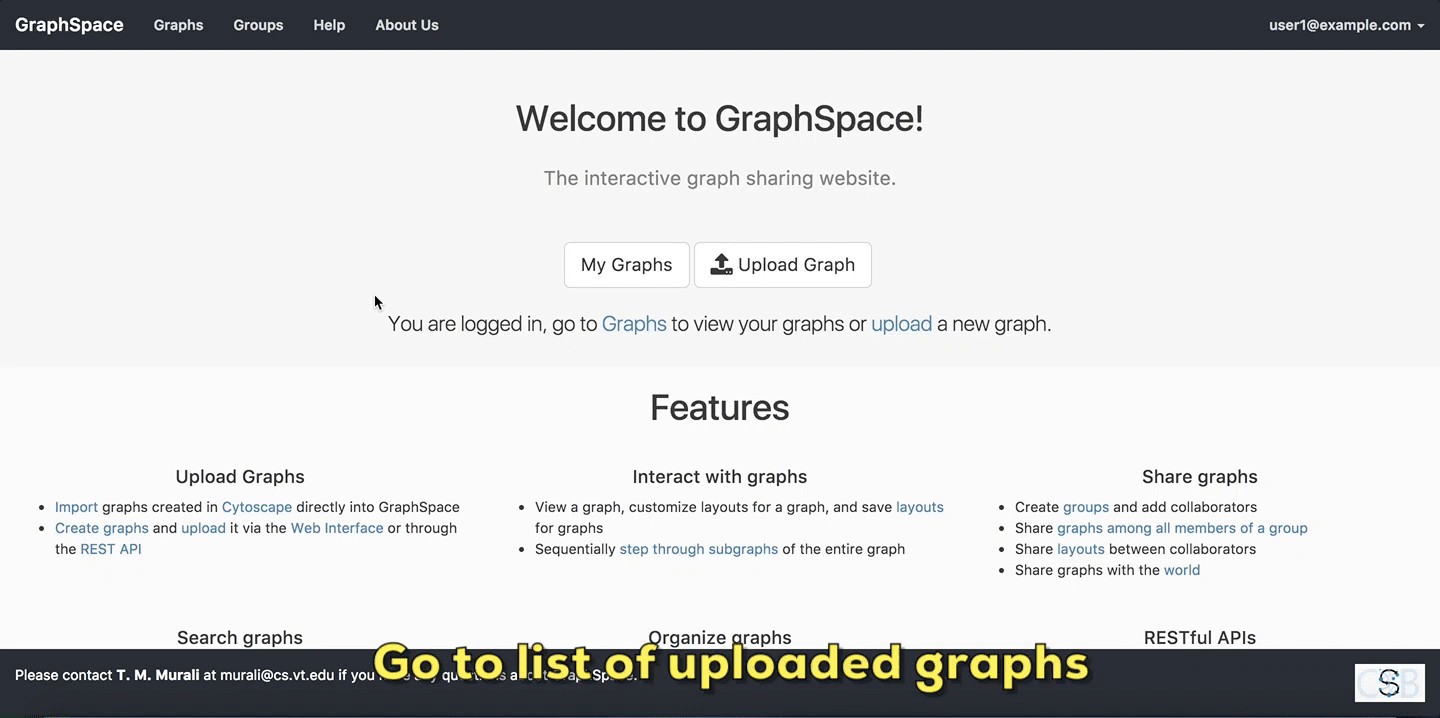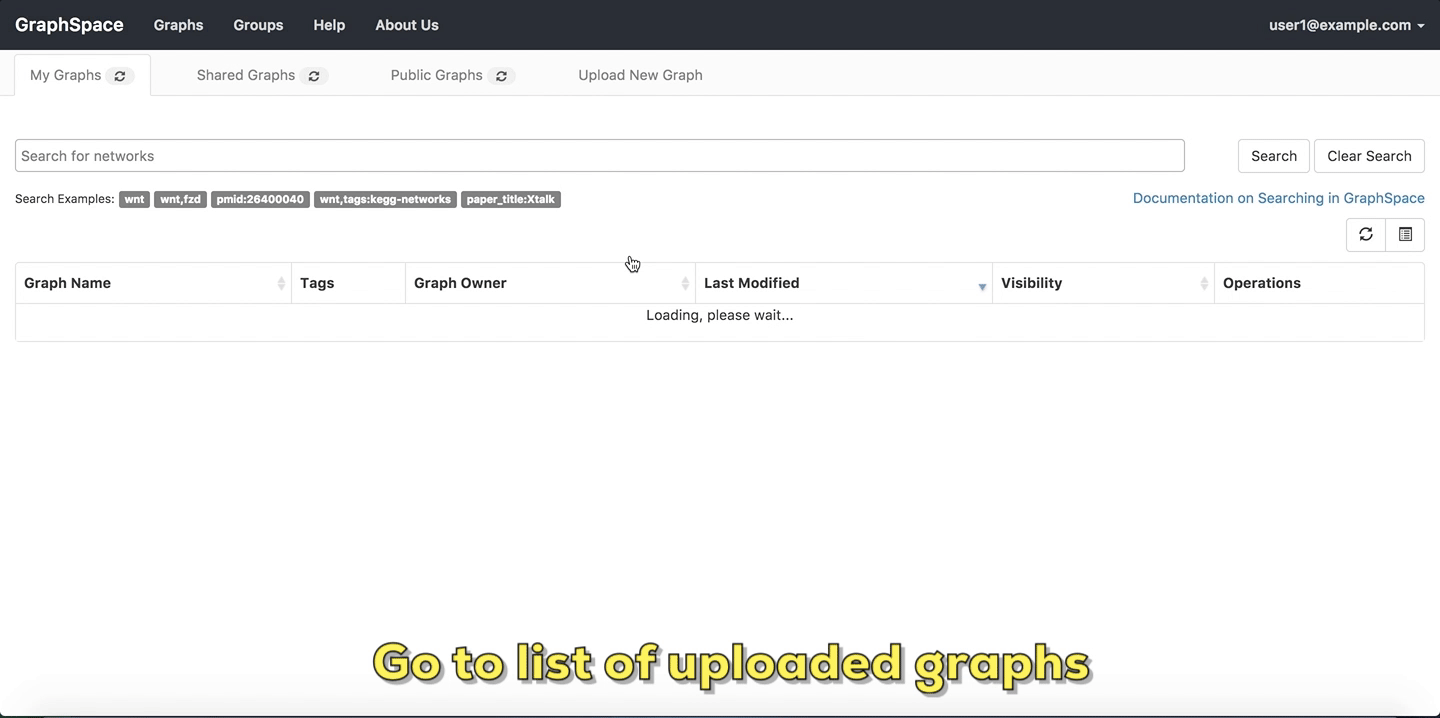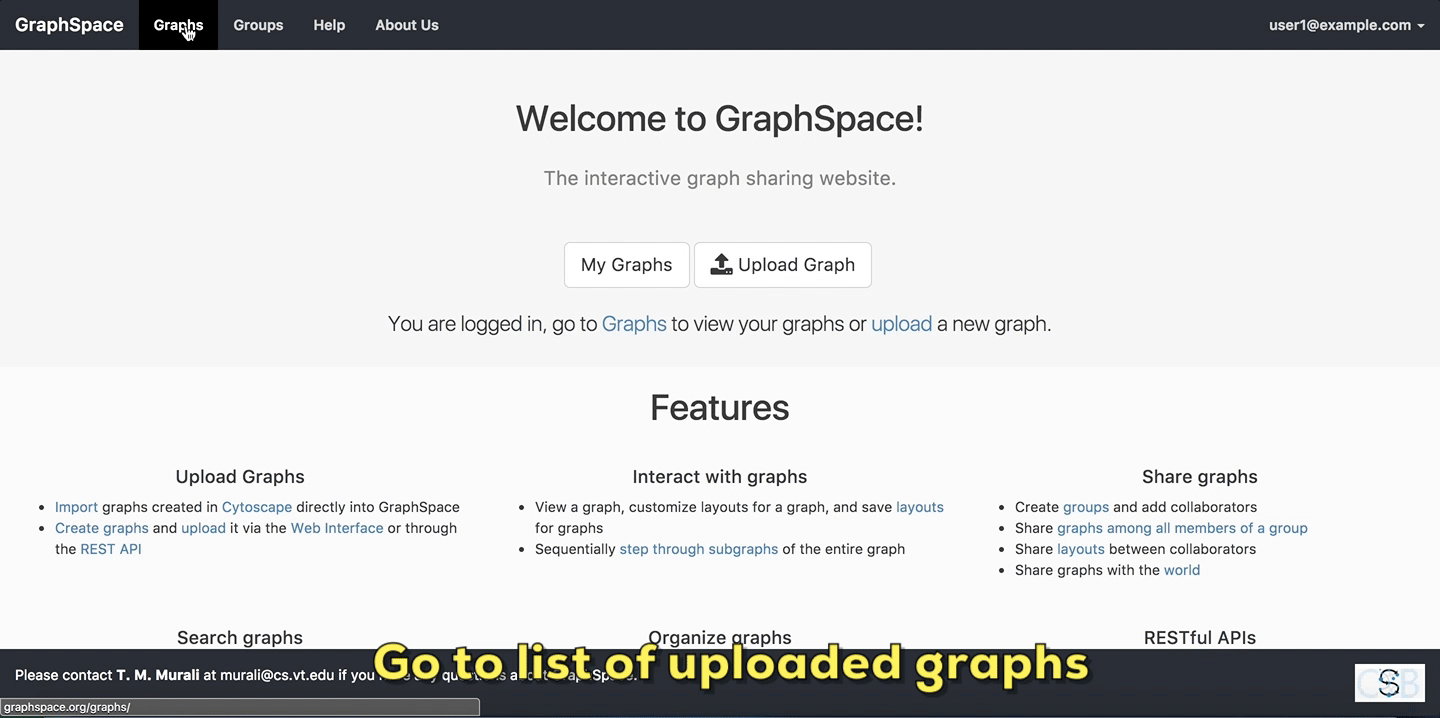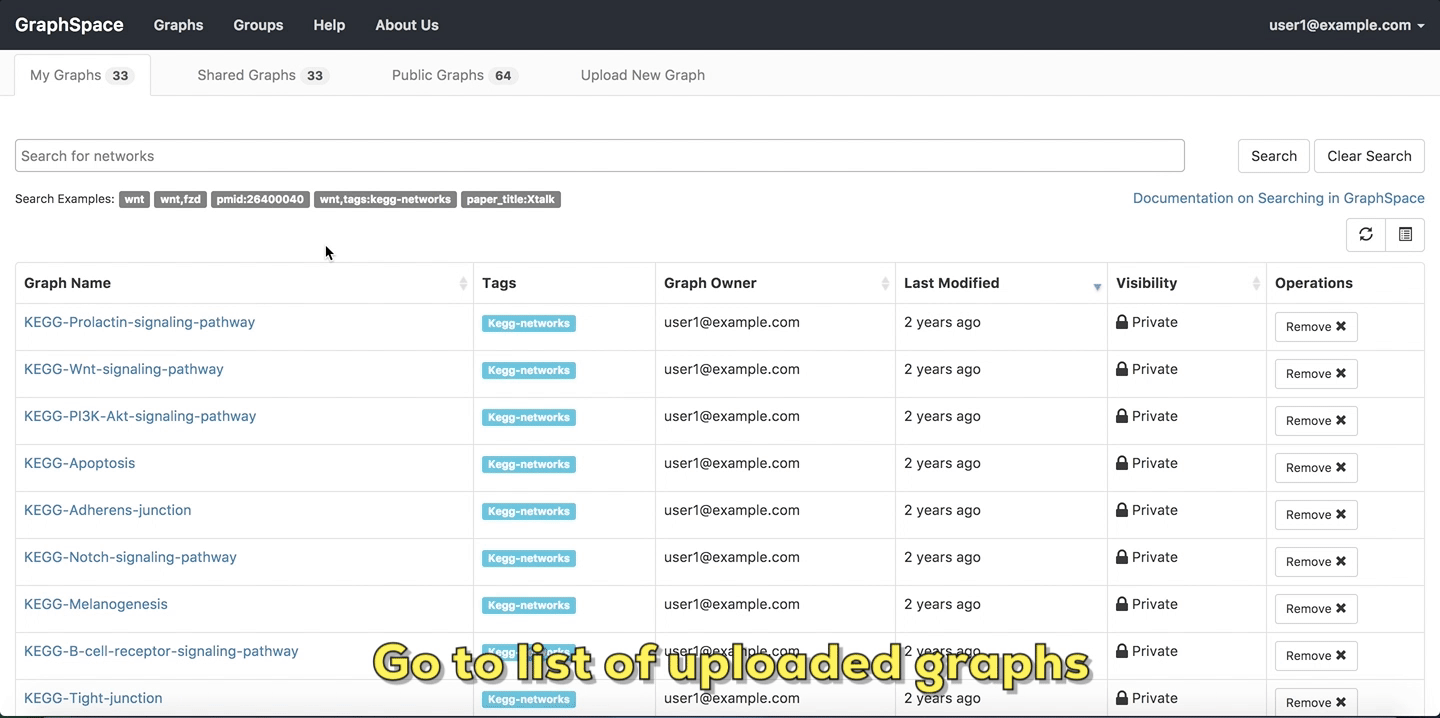 List of uploaded graphs
List of uploaded graphs
2.2. Searching within Multiple Graphs¶
The search interface in GraphSpace allows a user to search for networks that have a specific attribute or tag and contain one or more nodes using simple syntax on Graphs Page by following the given steps:
- Enter the name of the graph/node or specific network attribute mapping you are searching for in the search bar.
- Press
Enterkey or click on theSearchbutton.
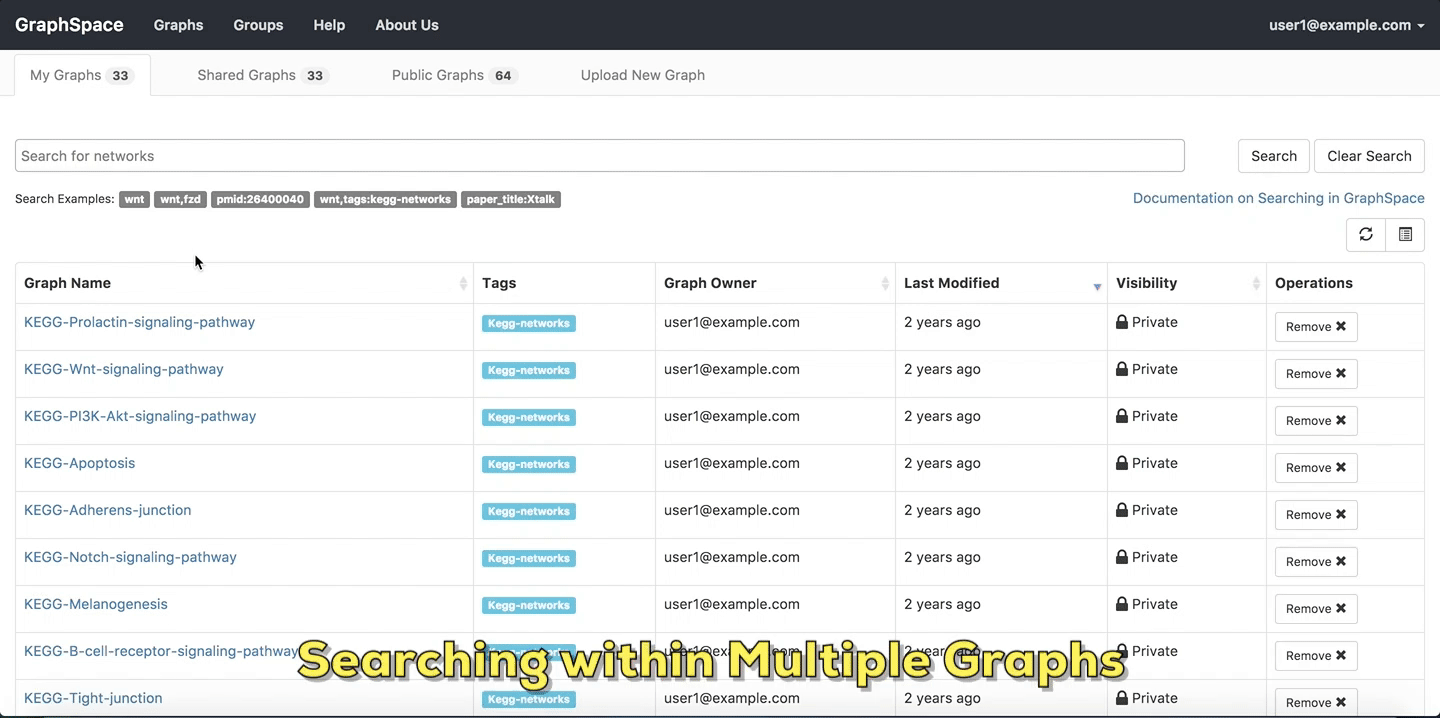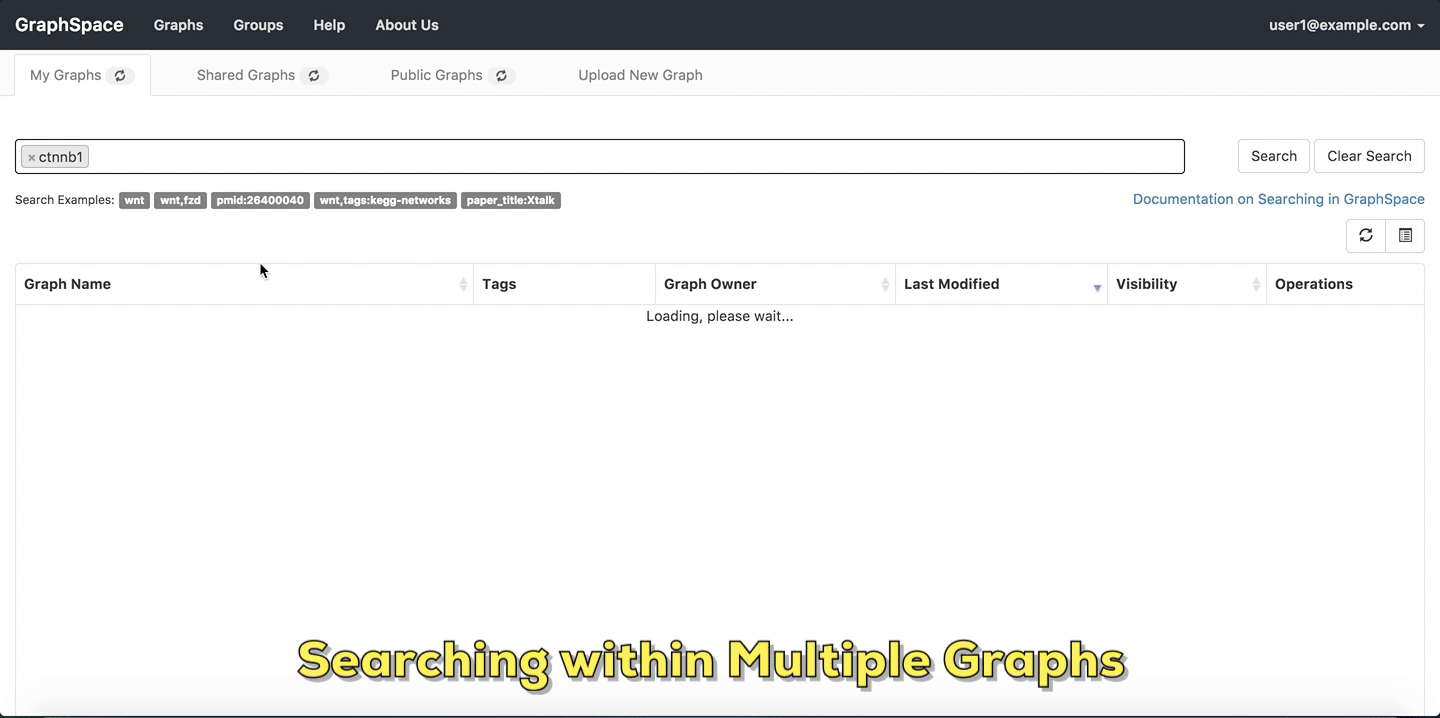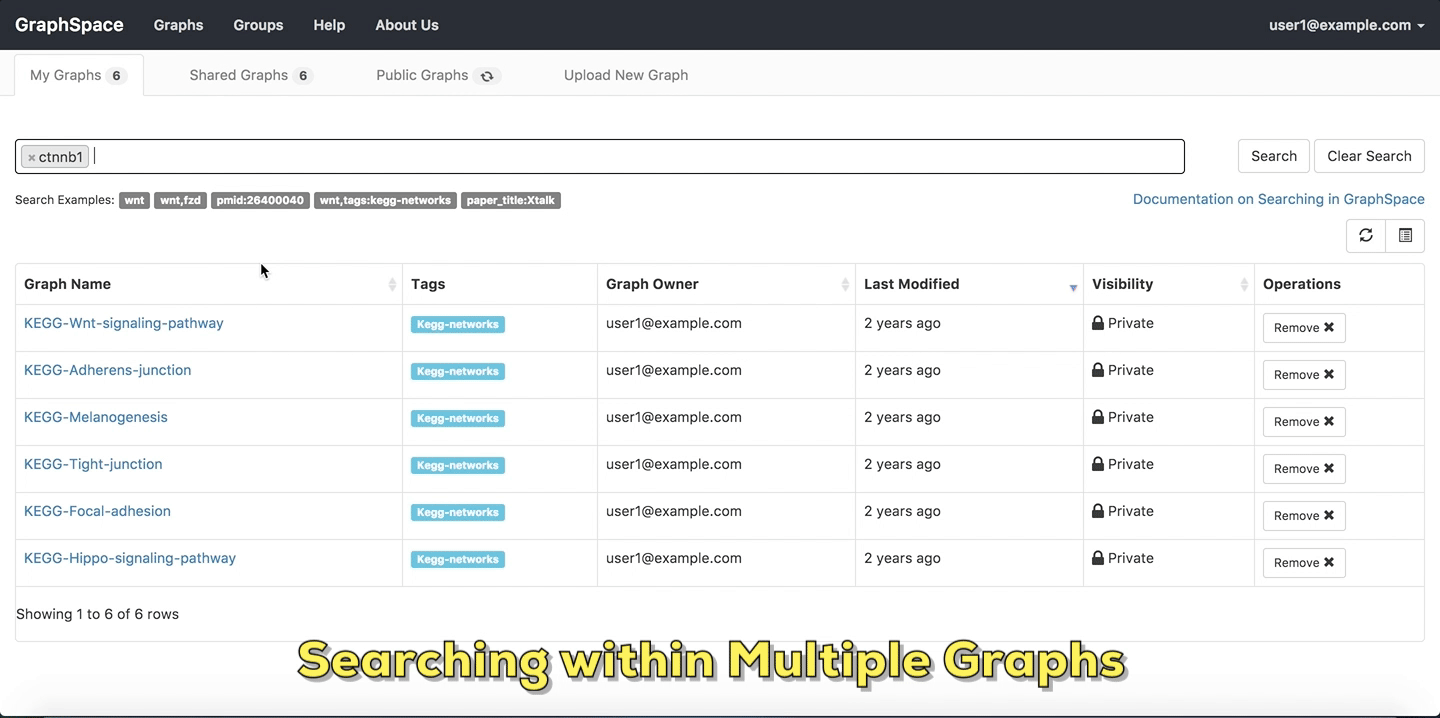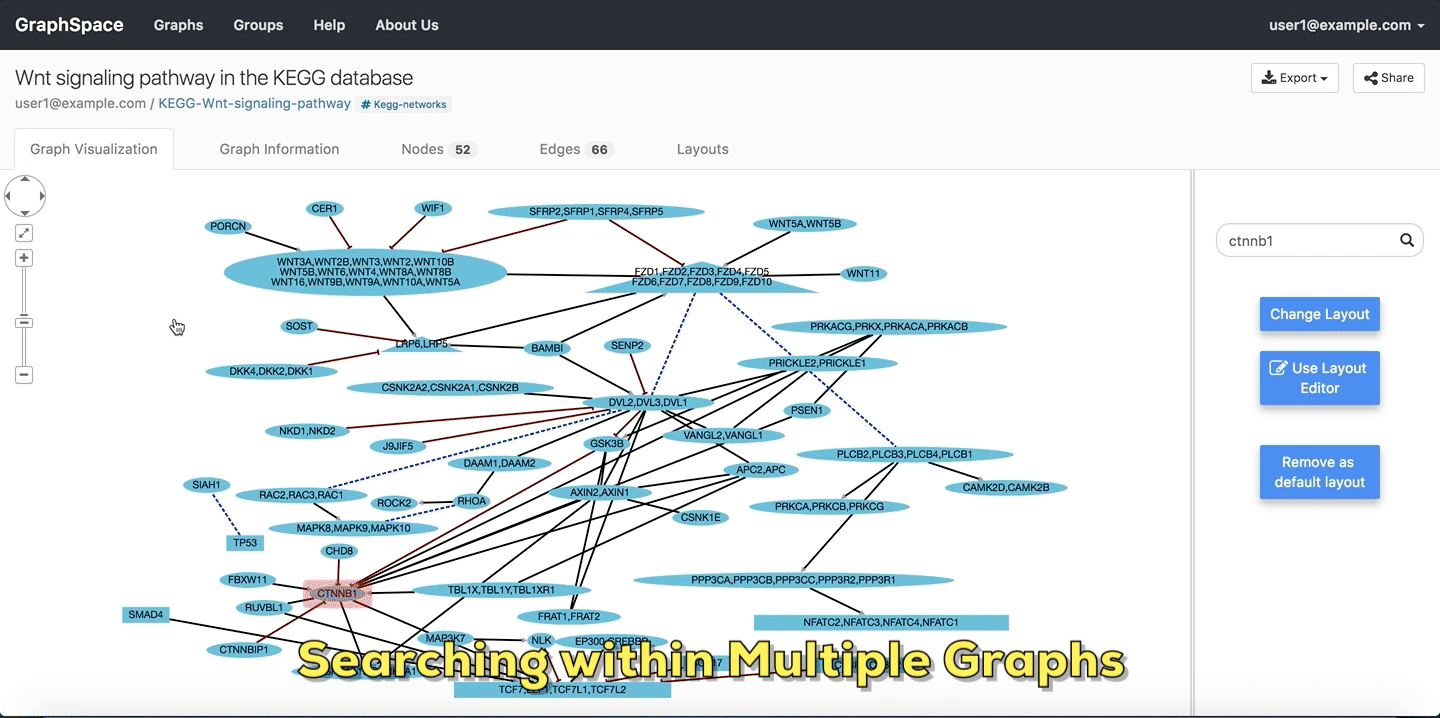
In this example, the user searches for the list for graphs that contain the protein (node) CTNNB1 (the symbol for β-catenin, a transcriptional regulator in the Wnt signaling pathway). The reduced list of graphs are the graphs where protein (node) name, label or aliases contain CTNNB1 as a substring. The match is case-insenstive. In the following example, There are six graphs owned by the user and thirty-two public graphs that contain this protein. Each link in the Graph Name column will take the user to a specific graph with the search term highlighted. In this example, the user clicks on the graph with the name KEGG-Wnt-signaling-pathway and reaches the graph for the Wnt pathway with the searched node highlighted.
 Searching within Multiple Graphs
Searching within Multiple Graphs
2.3. Searching within a Single Graph¶
The user can search for node or edges within a given graph on GraphSpace by following the given steps:
- Enter the name of the node or an edge you are searching for in the search bar.
- The nodes or edges are highlighted automatically as you type in the name of the node or edge in the search bar.
In the following example, the user searches for the graph for two proteins (nodes) CTNNB1 and WNT using the query ctnnb1, wnt. This search query highlights the proteins where protein (node) name, label or aliases contains CTNNB1 or WNT as a substring (case-insensitive). In the following example, the graph contains four nodes which match the given query.
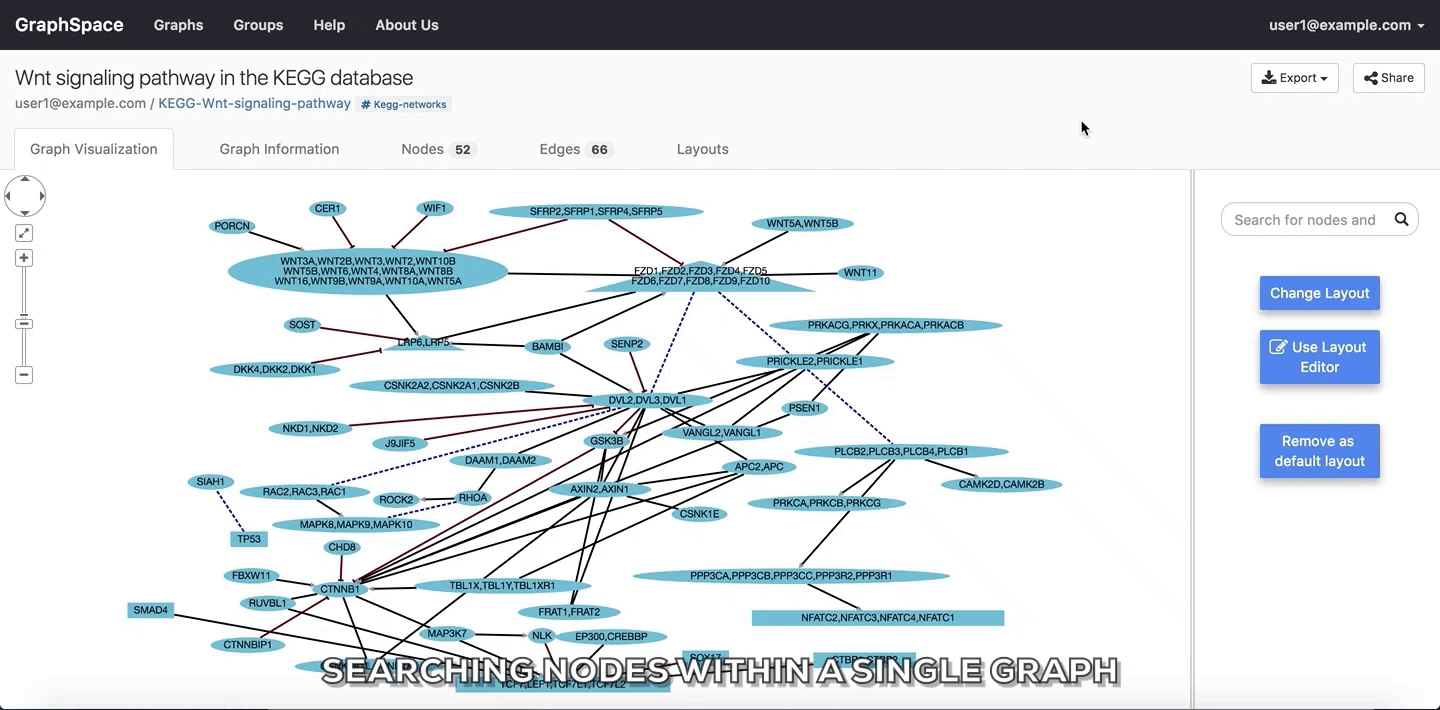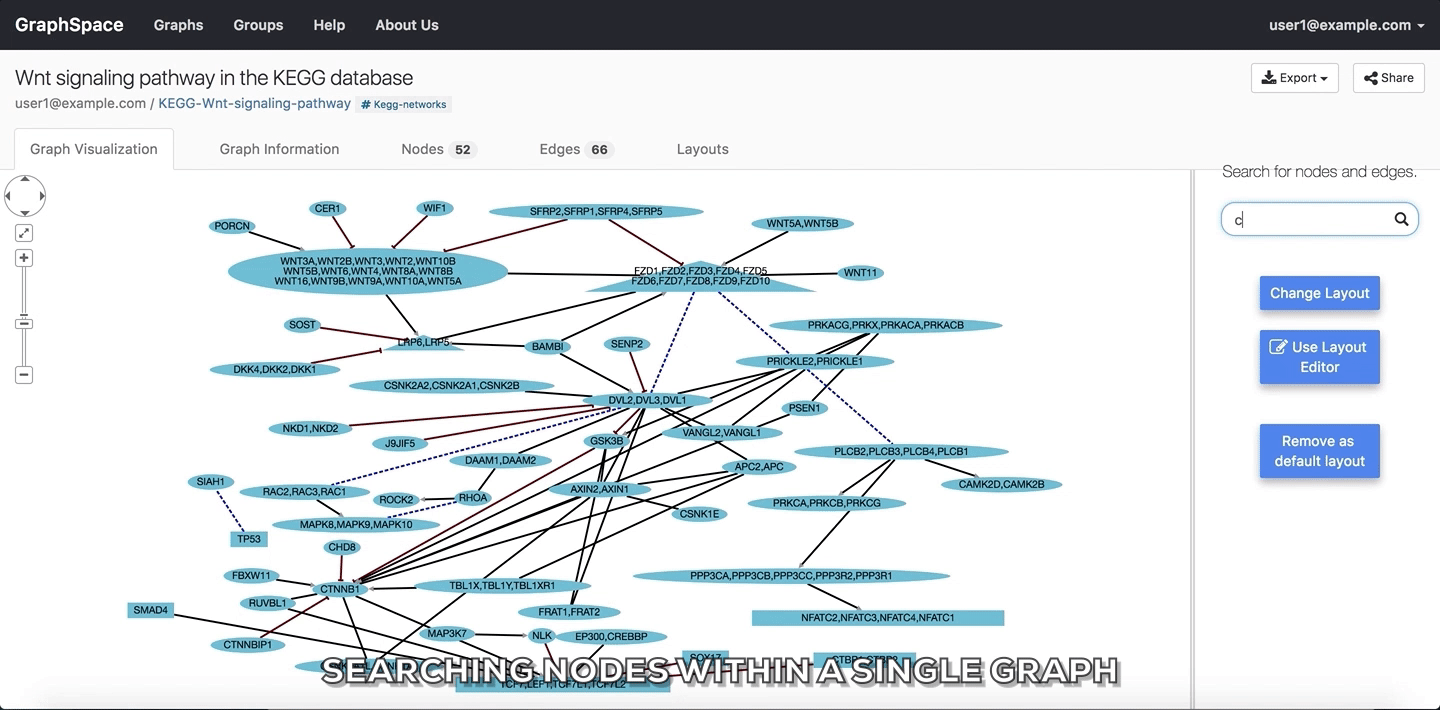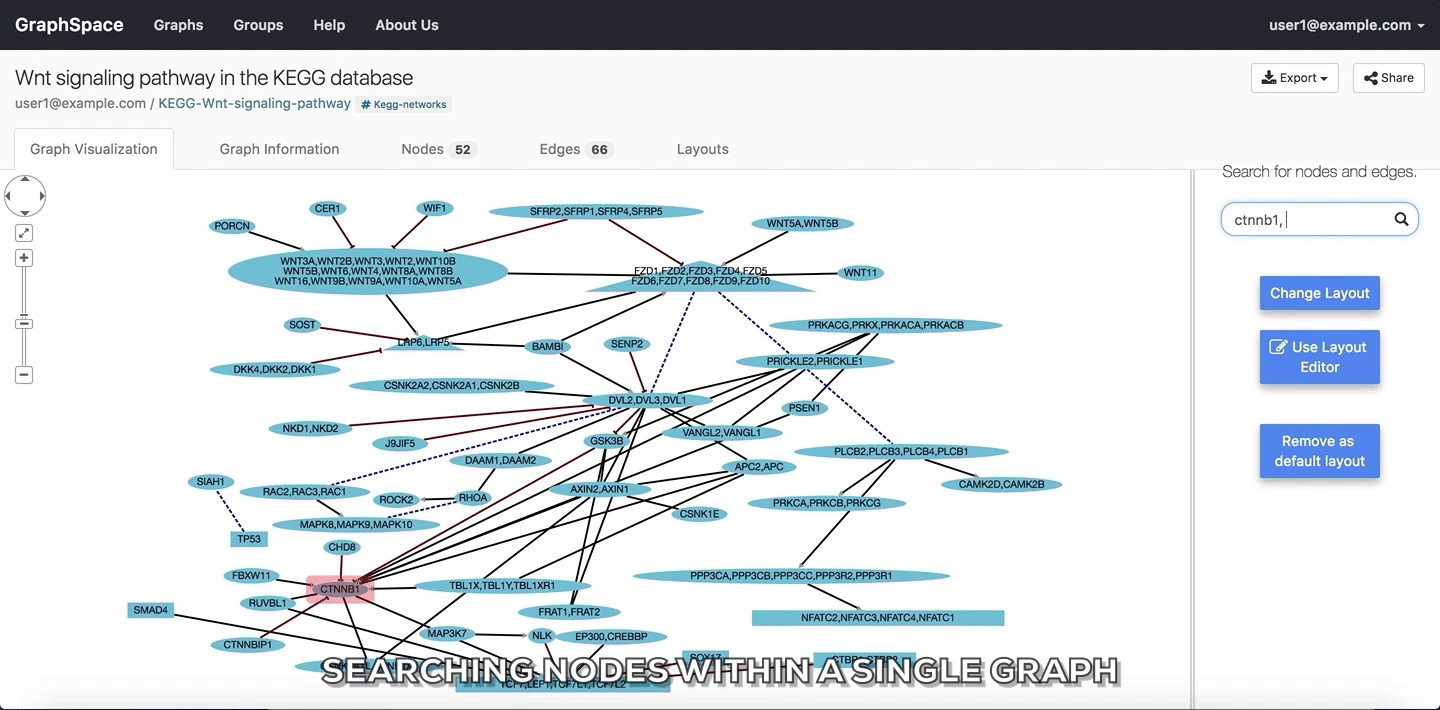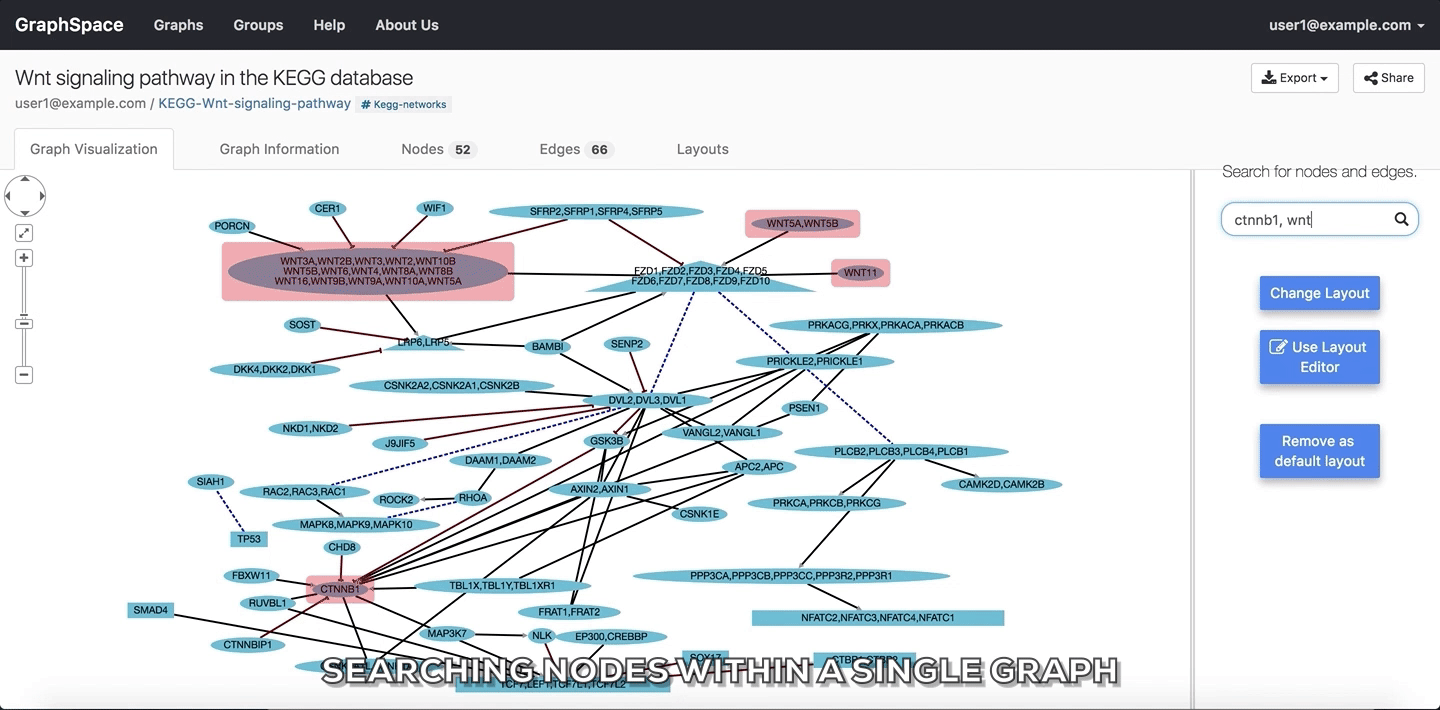 Searching nodes within a Single Graph
Searching nodes within a Single Graph
In the following example, the user searches the graph for edges from Wnt to Fzd using the query Wnt::Fzd. GraphSpace
highlights any edge whose tail node matches wnt and whose head node matches fzd. In the following example, the graph contains three edges which match the given query.
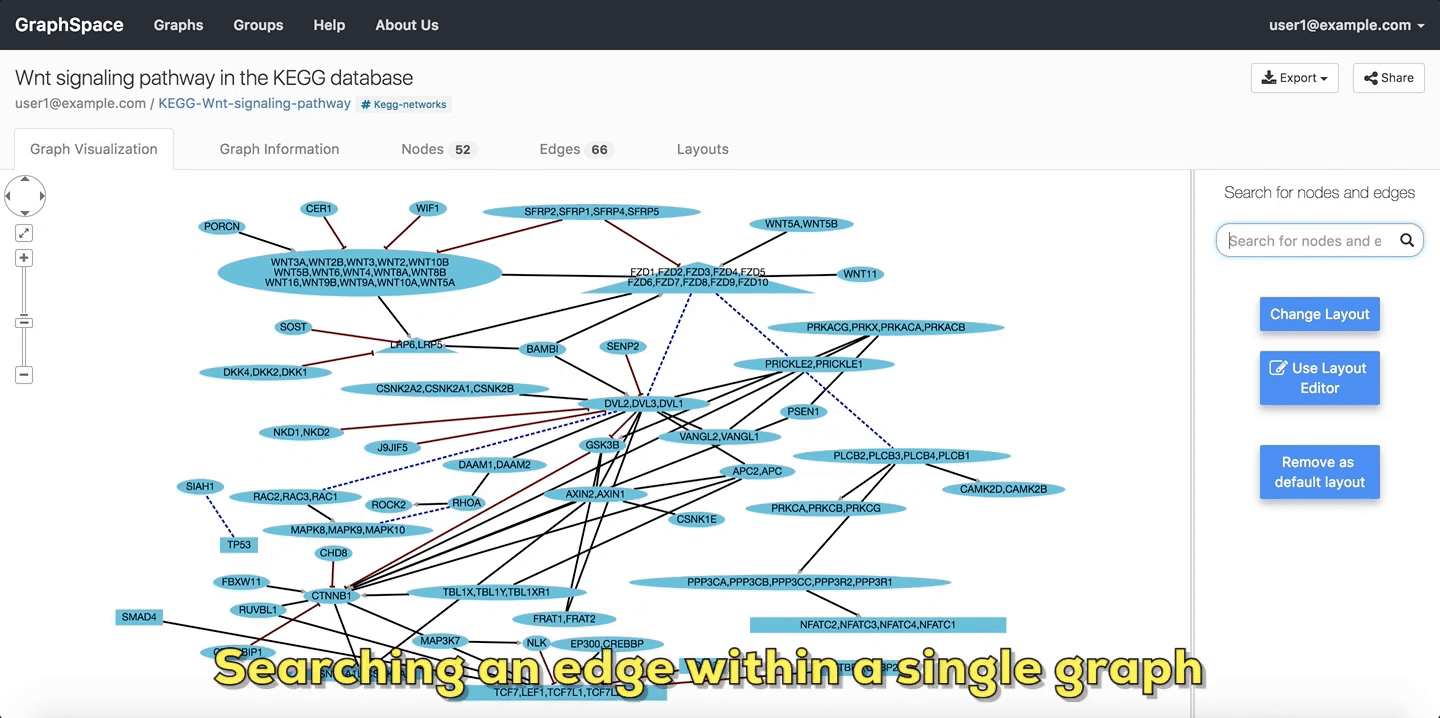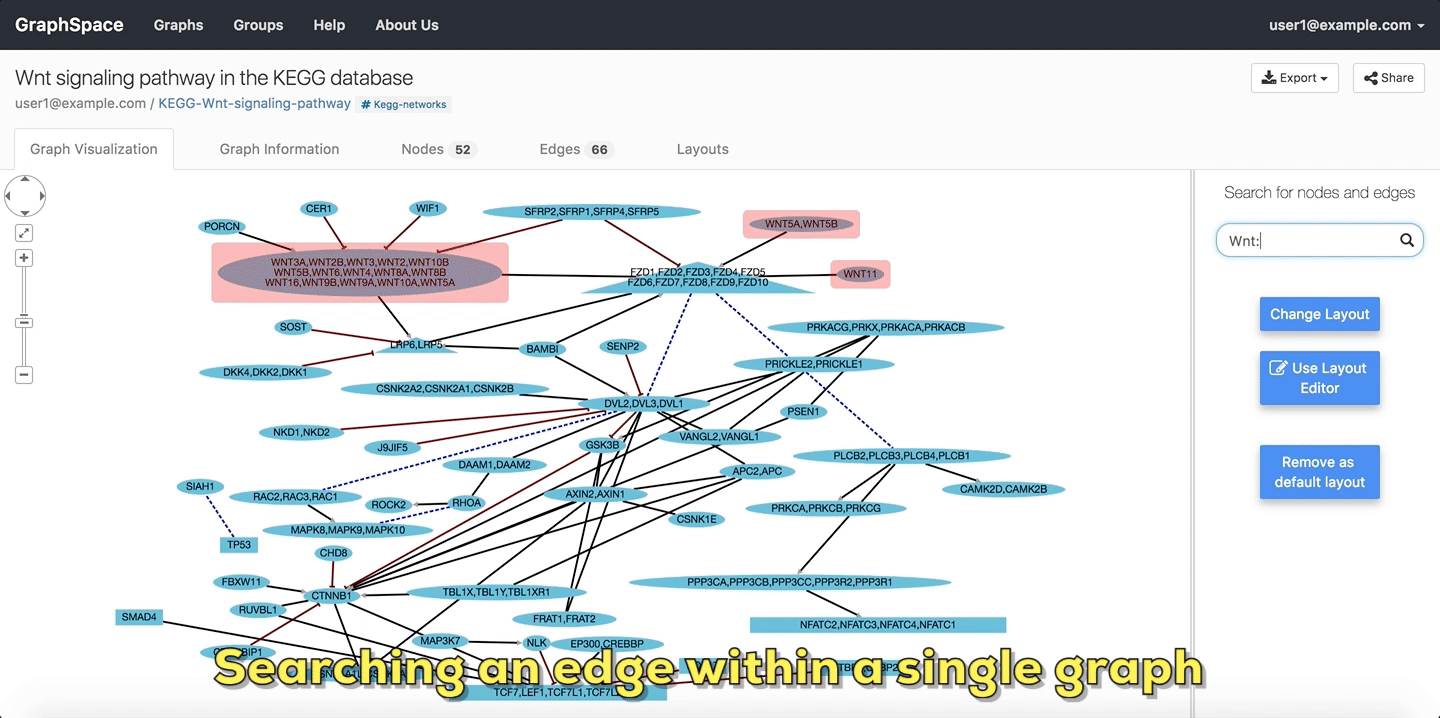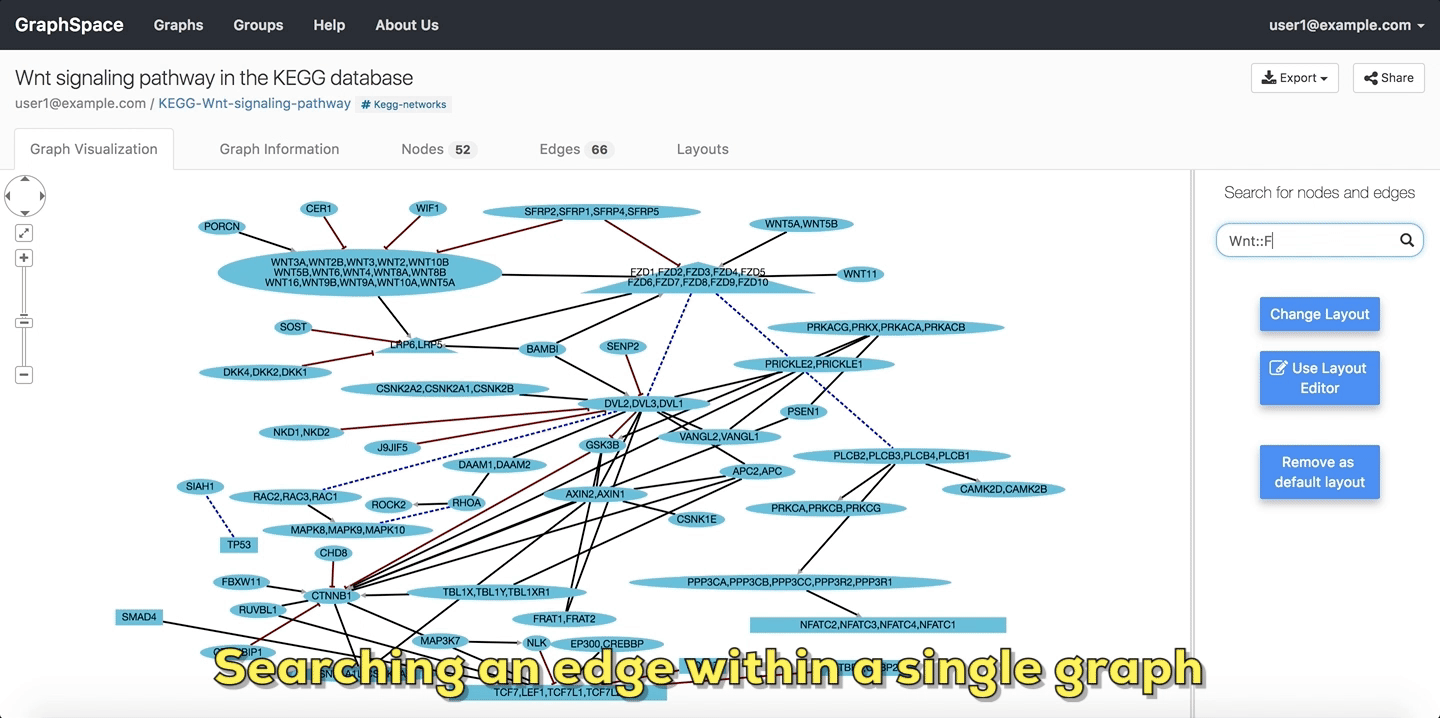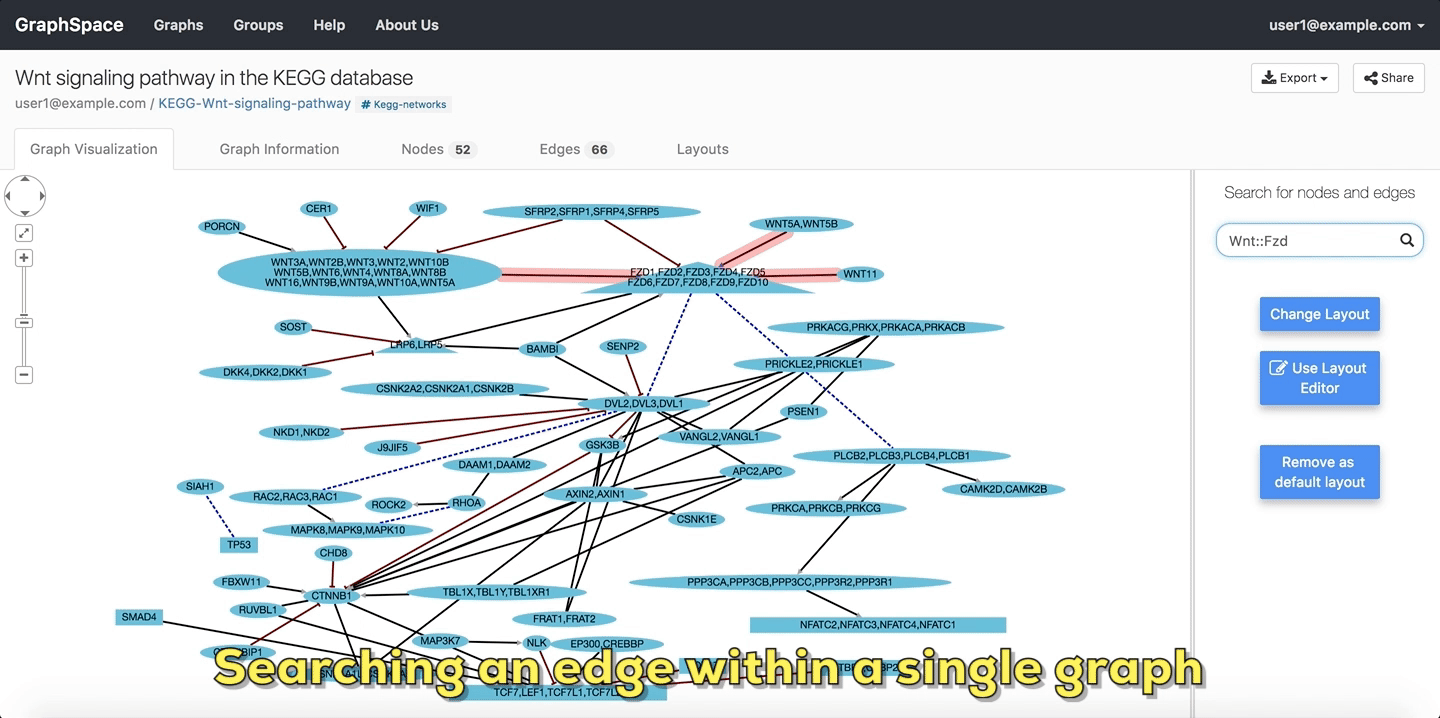 Searching edges within a Single Graph
Searching edges within a Single Graph
2.4. Interacting with a Graph¶
In this section, we examine different ways to interact with an individual network on its page. The information that appears in following examples must be included in the JSON files that are uploaded by the network owner, as described in the Network Model section. An individual network page is designed to access features like:
2.4.1. Graph Information¶
As its name suggests, the Graph Information panel displays information about the entire graph, e.g., a legend of node and edge shapes and colors. The description attribute in the JSON for the network specifies this content. The user can go to Graph Information panel by clicking on the Graph Information link above the graph.
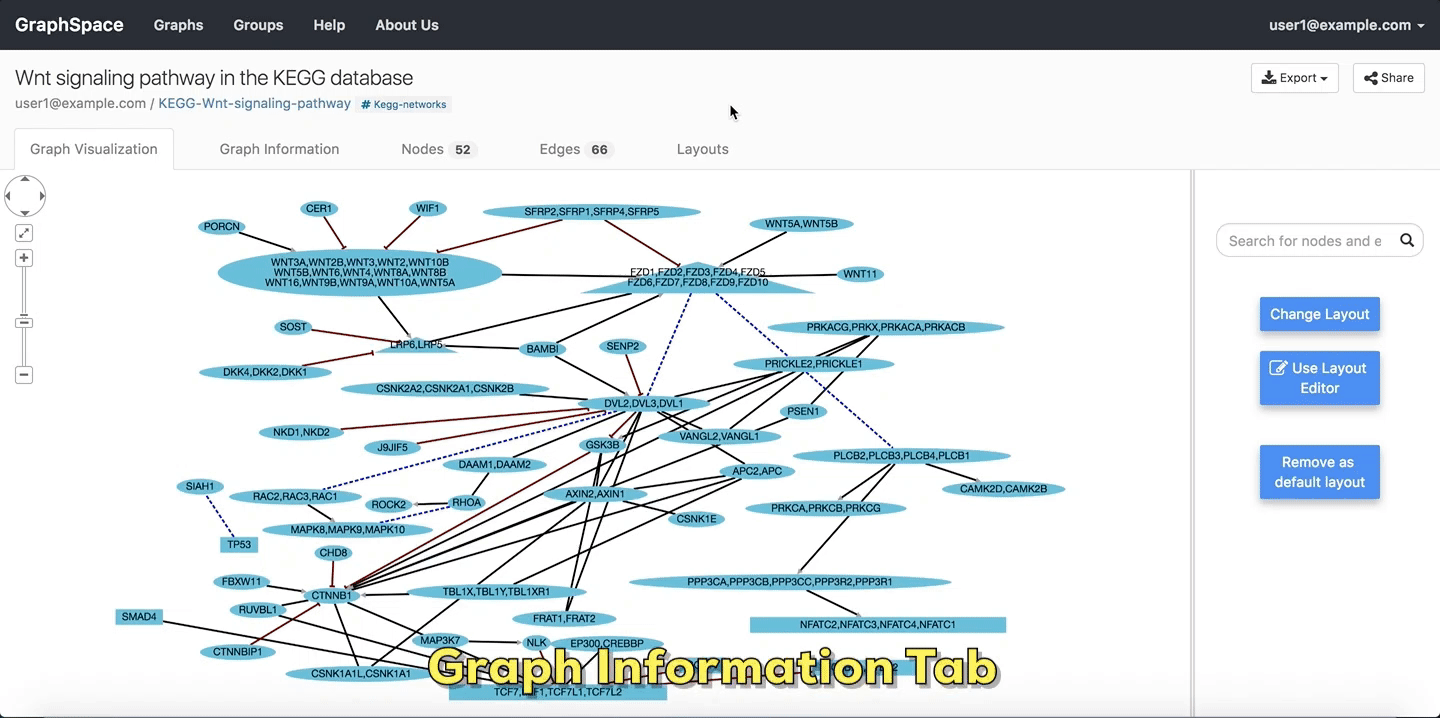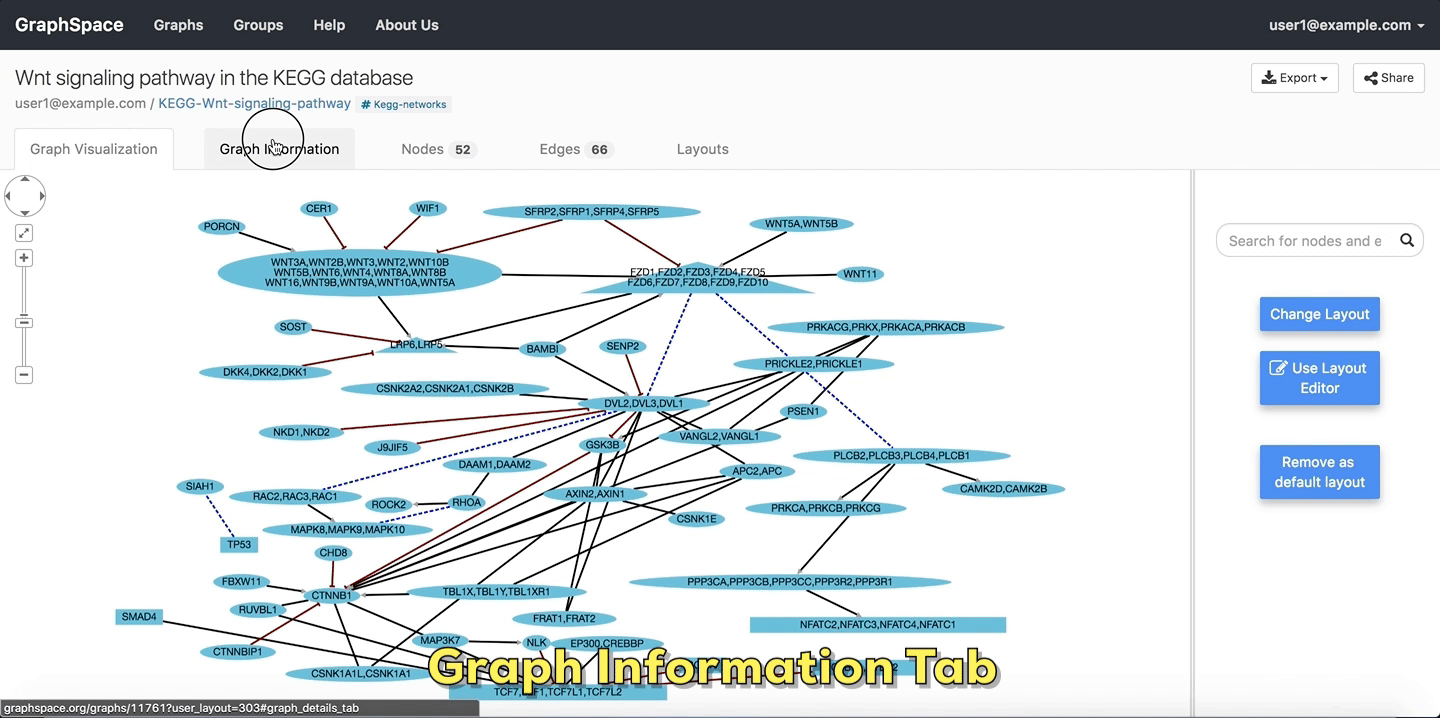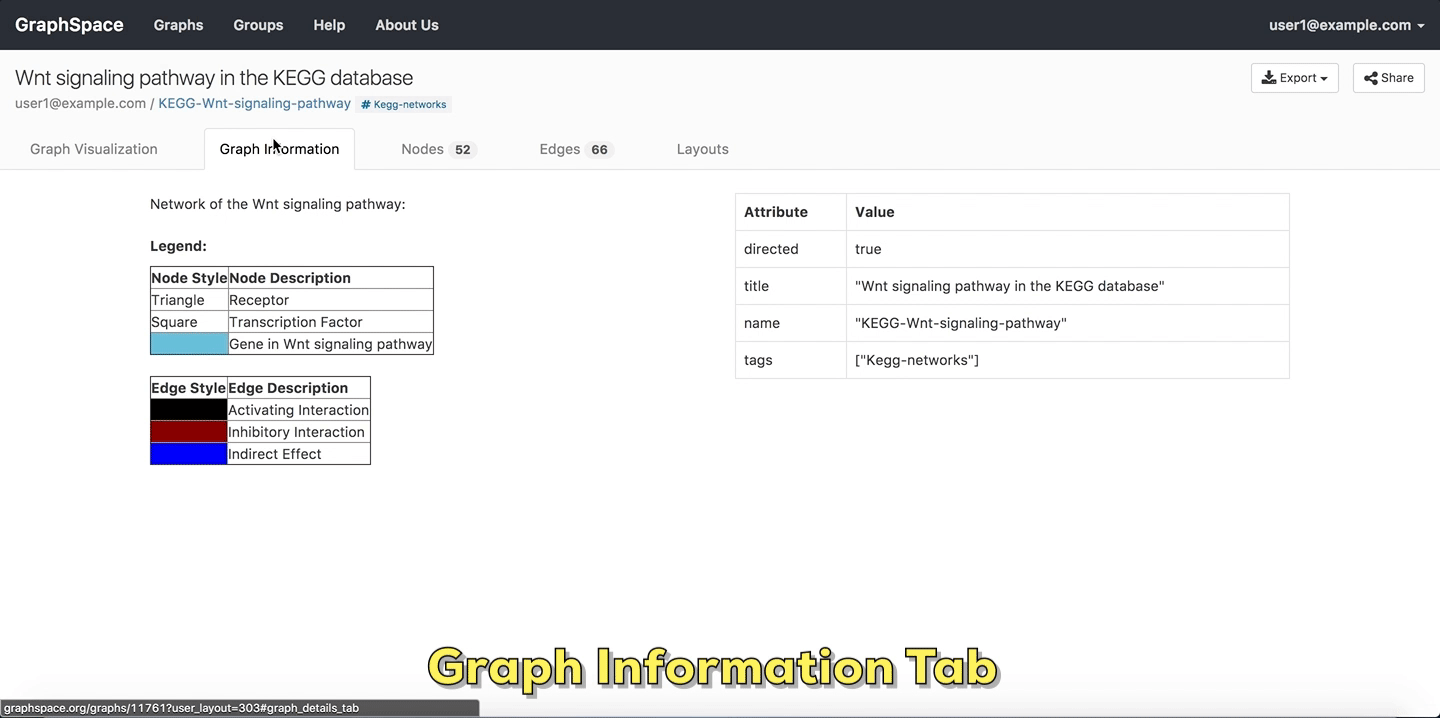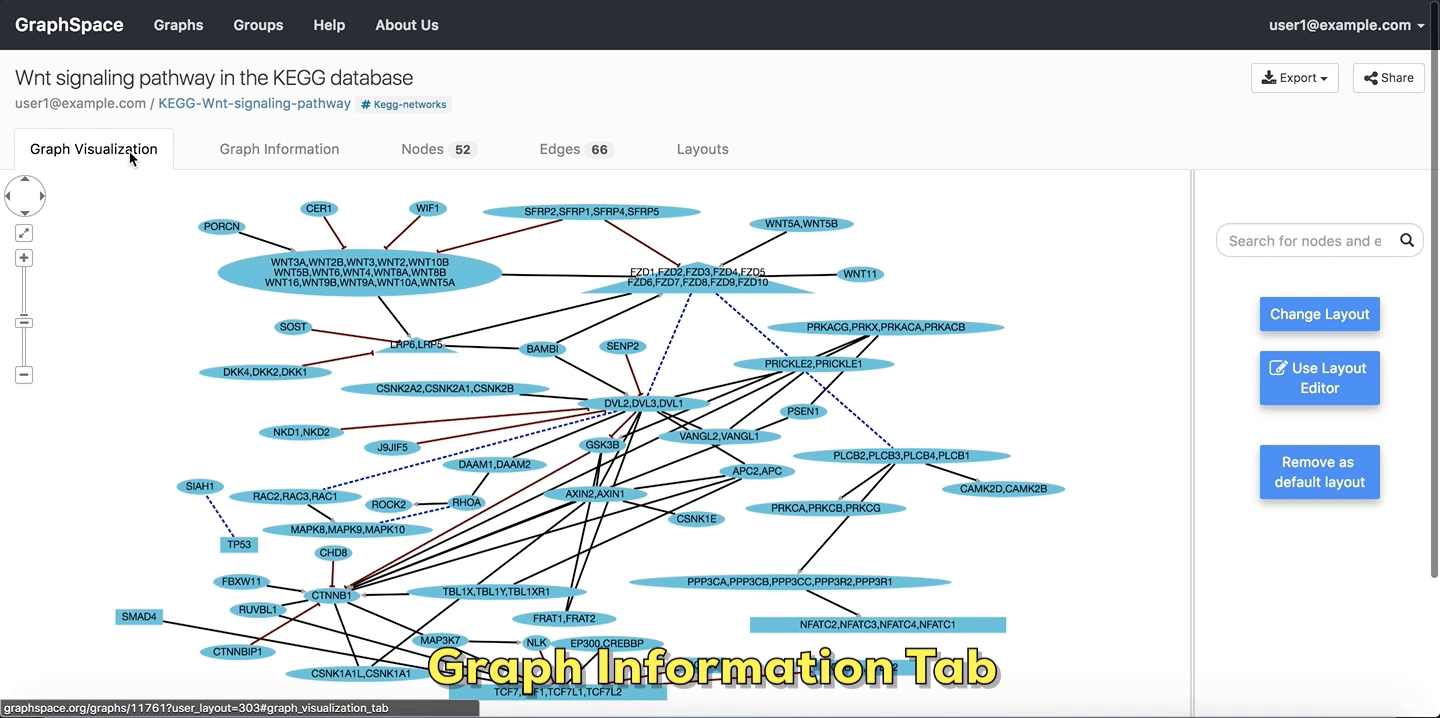 Graph Details
Graph Details
Please refer to this section for more information.
2.4.2. Edge and Node Information¶
Clicking on a node or edge pops up a panel with information on that node or edge. The popup attribute for the node in the network JSON specifies this content.
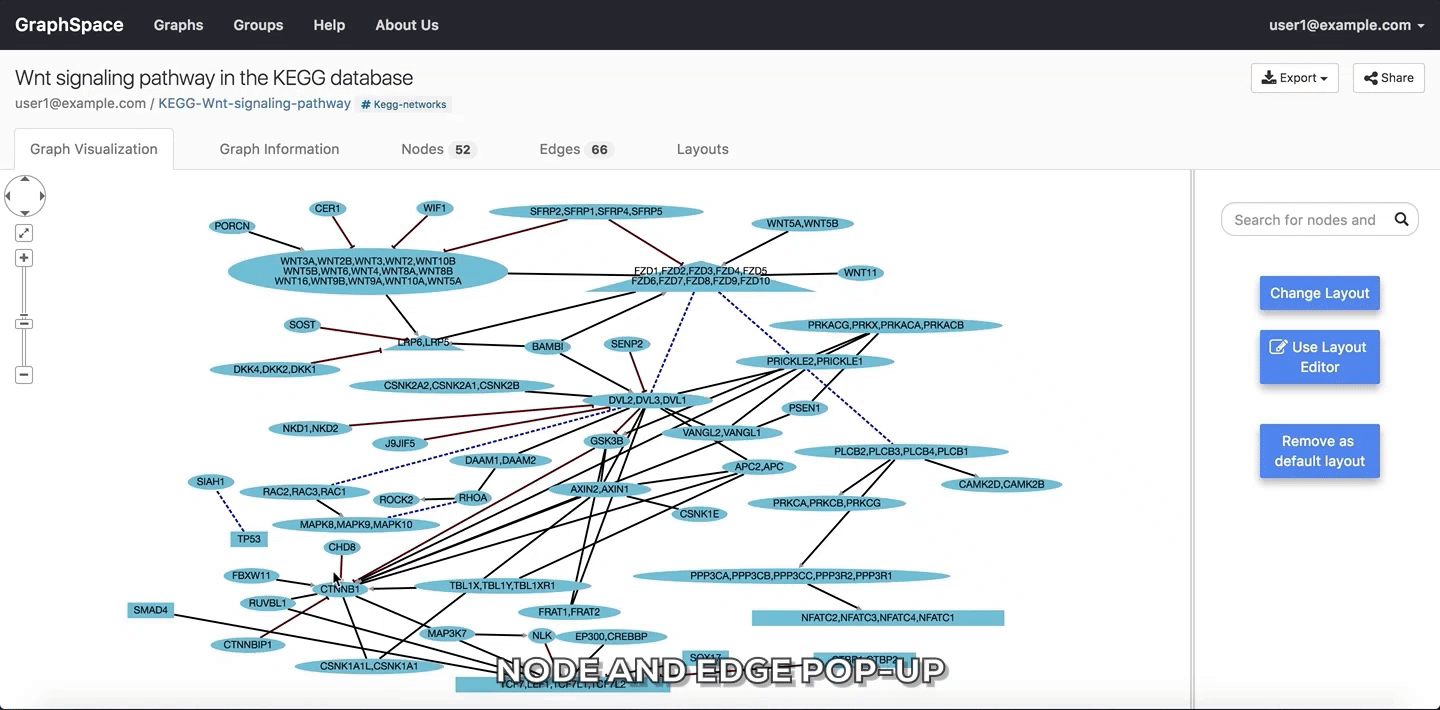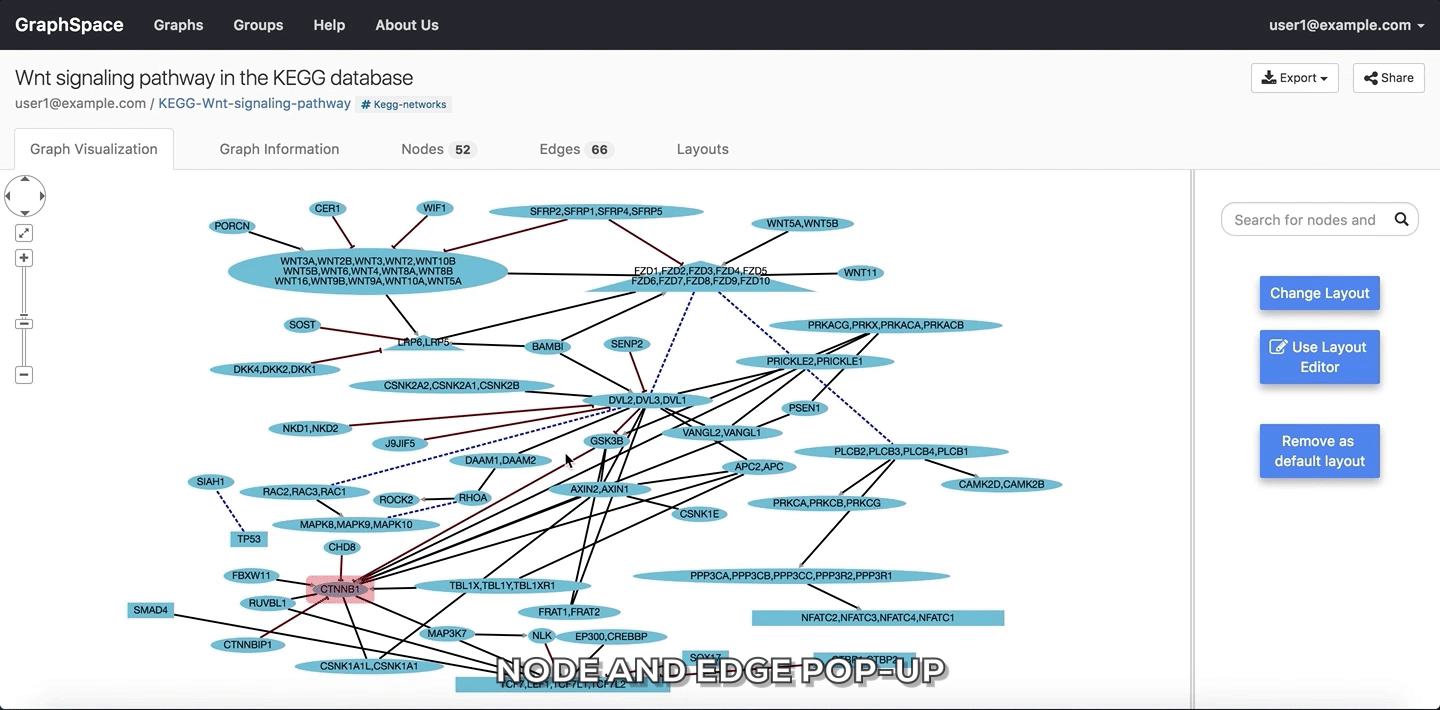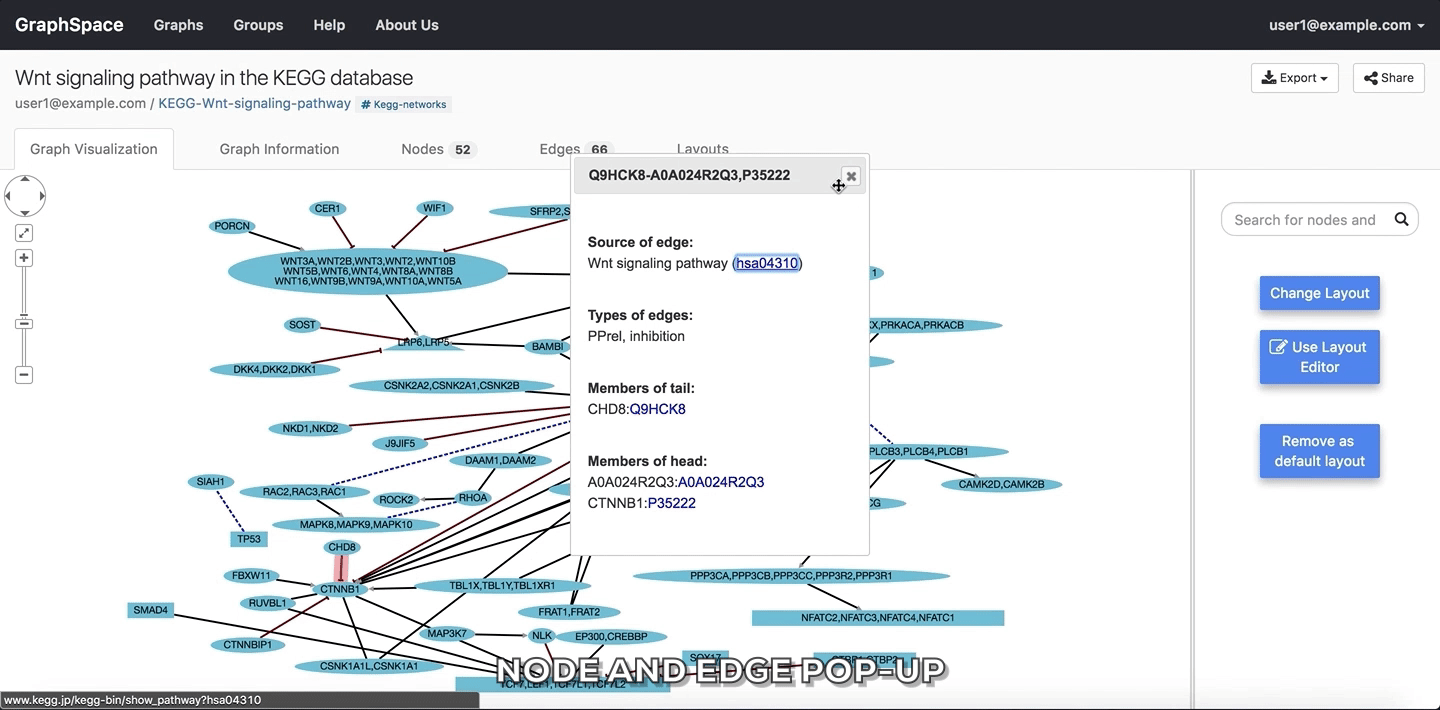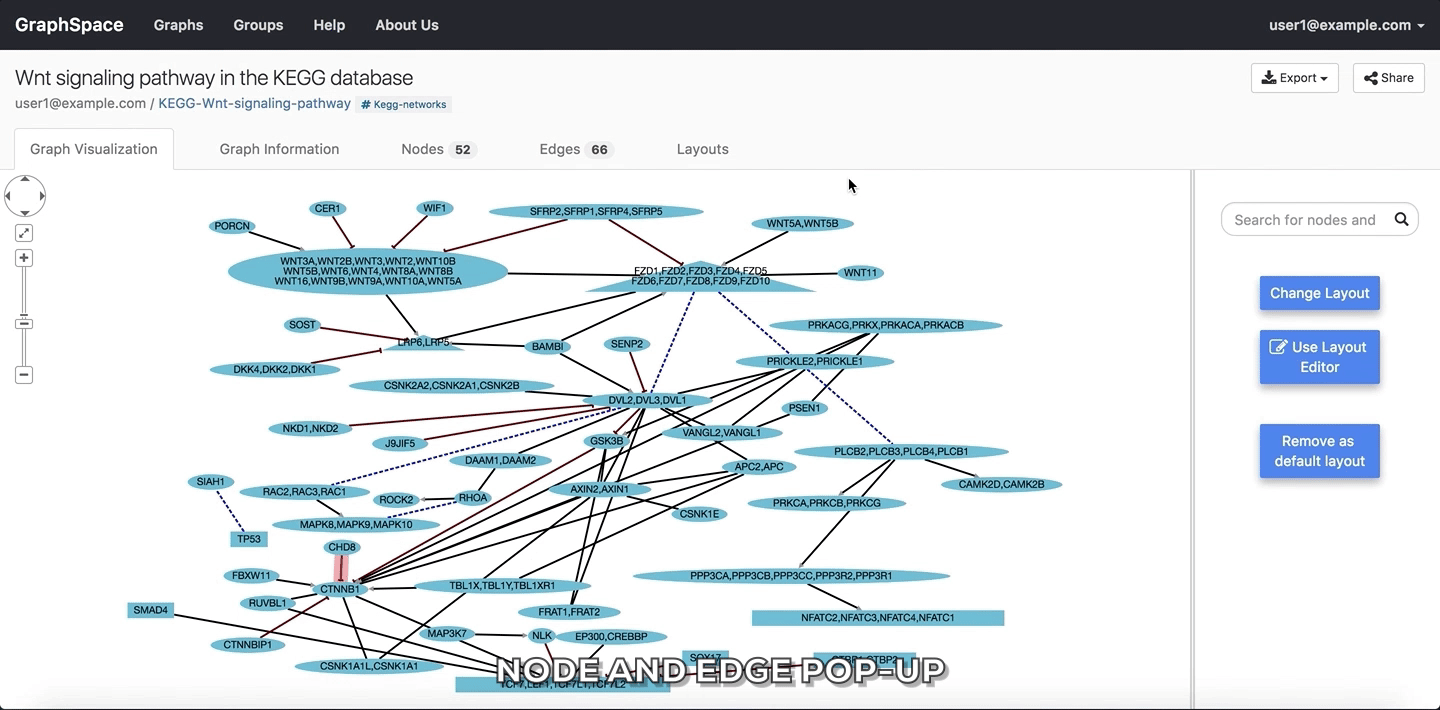 Edge and Node Information
Edge and Node Information
Please refer to this section for more information.
2.4.3. Export Graph¶
GraphSpace allows users to export a graph as an image file or a JSON file. GraphSpace does not support any other export formats since it relies on Cytoscape.js for this functionality, which implements only export to PNG, JPG and JSON format.
In the following example, the user is exporting the graph as an image in PNG format.
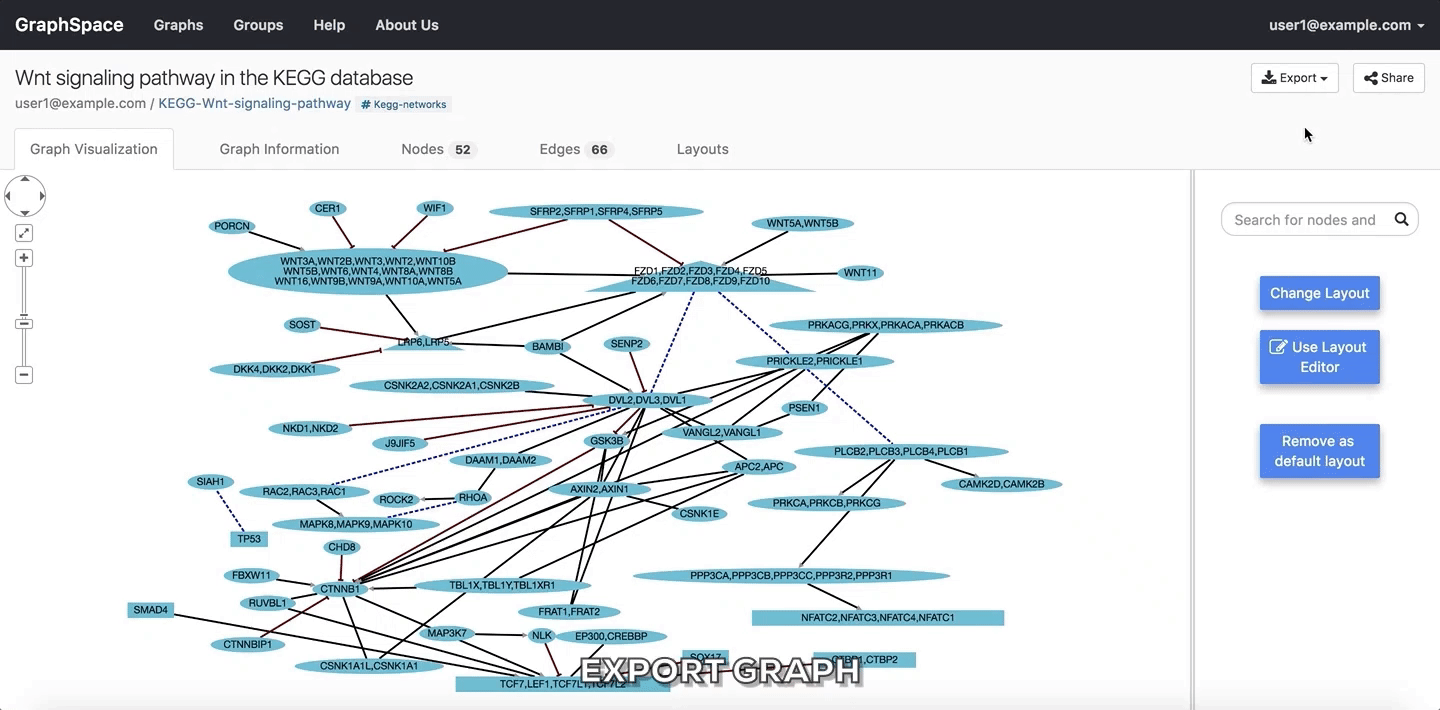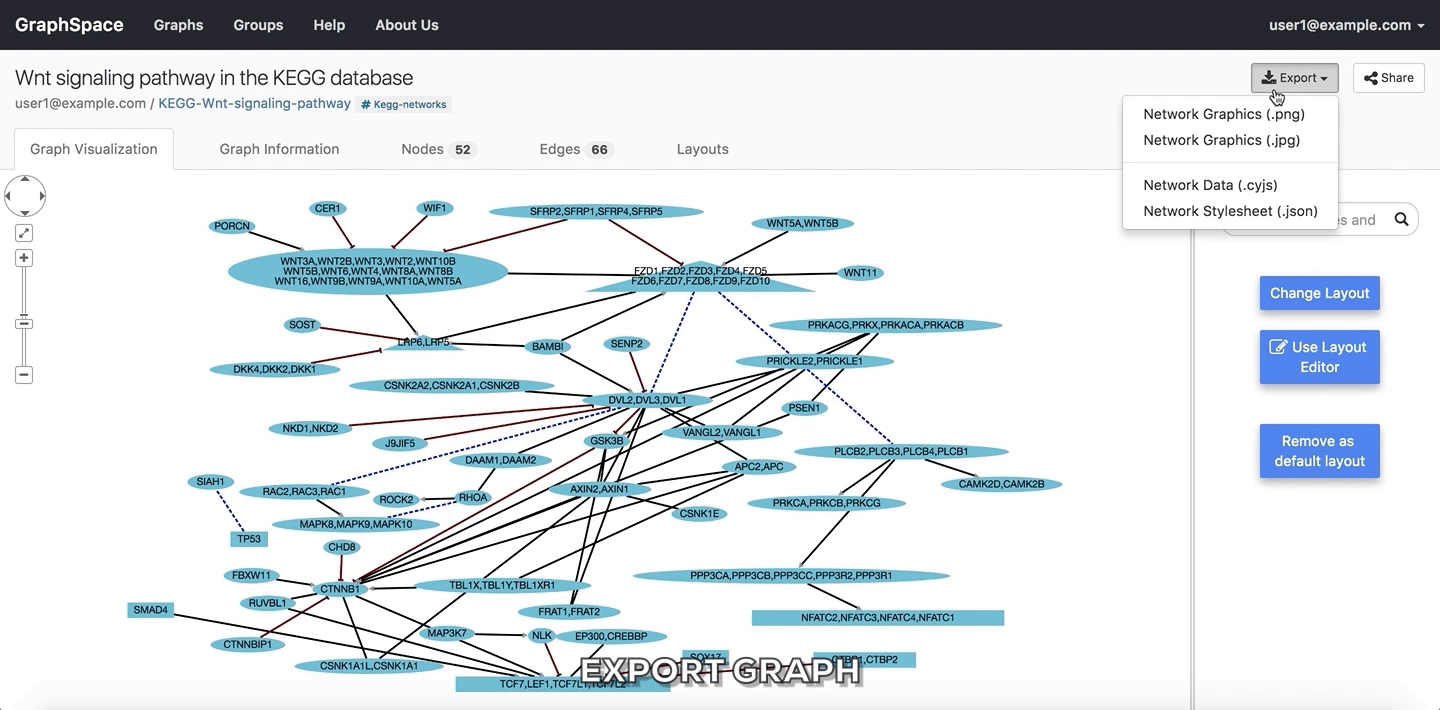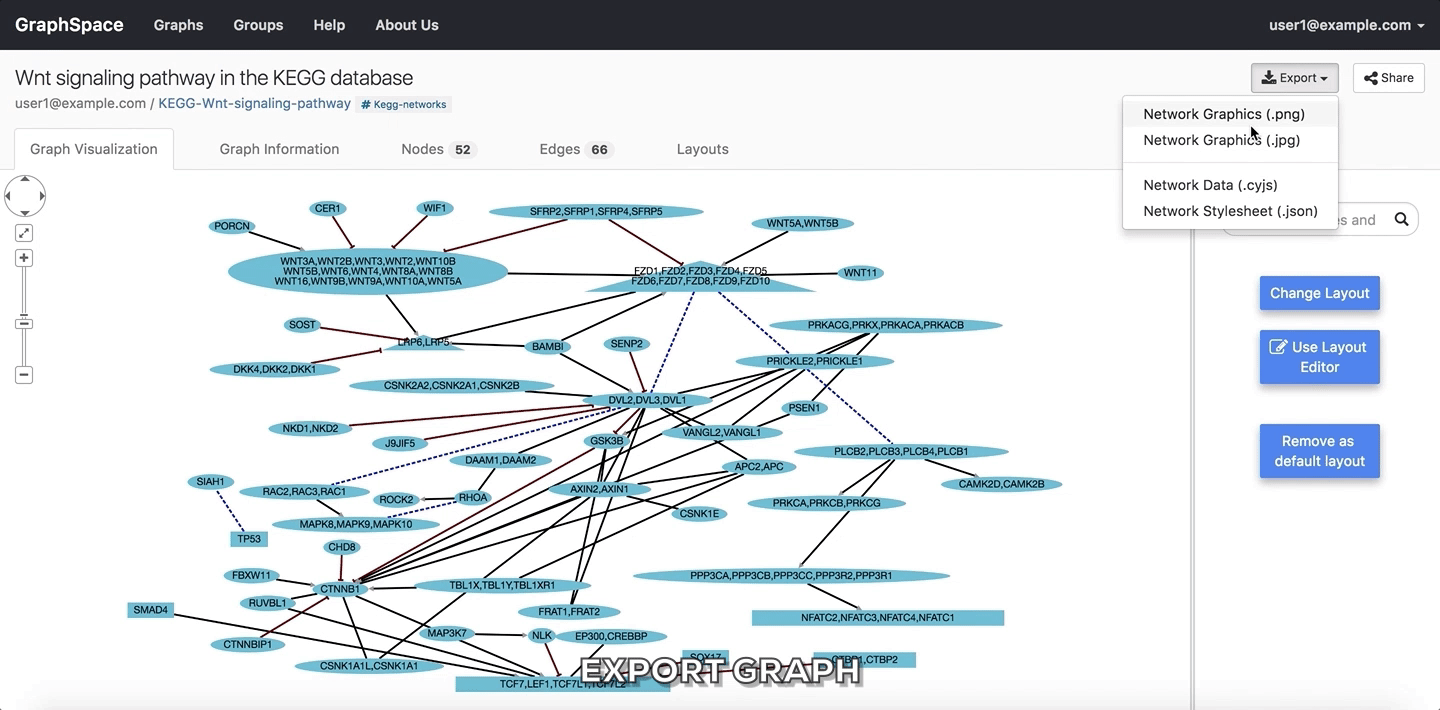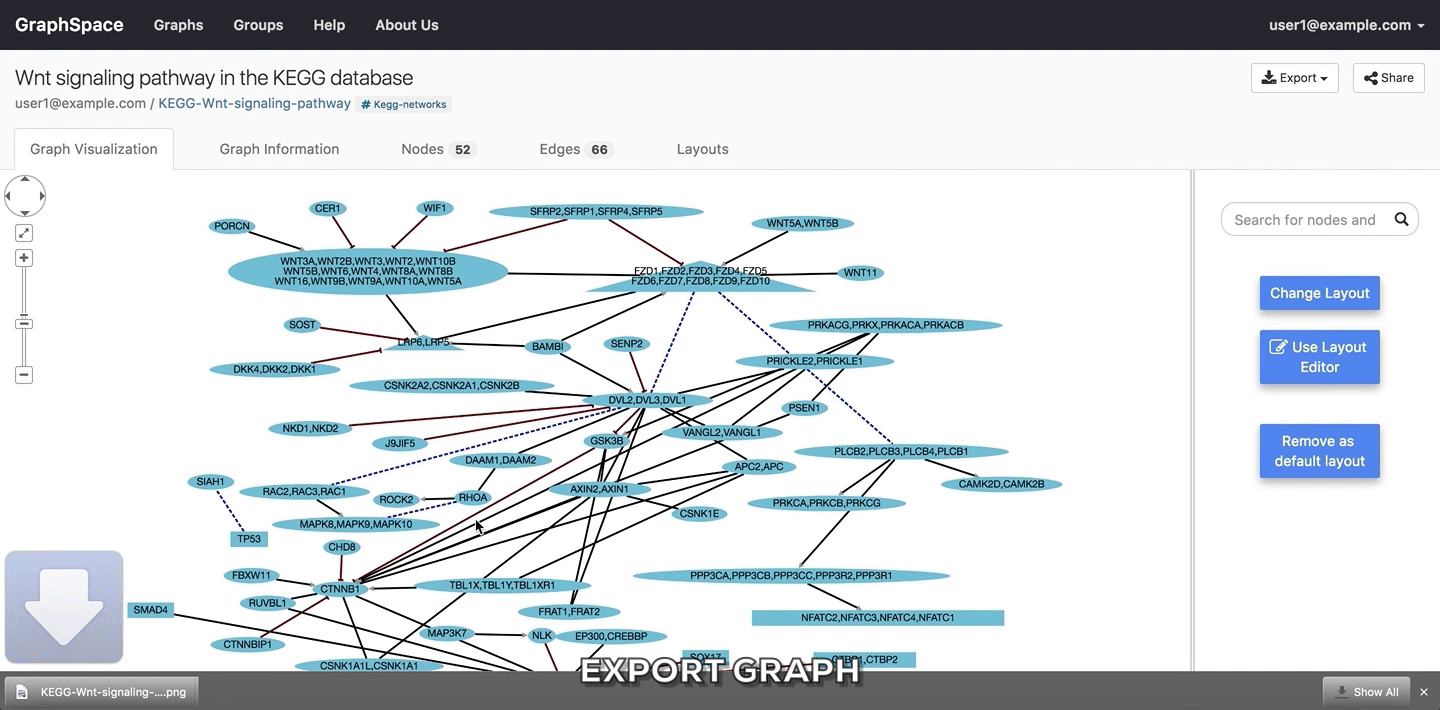 Export Graph
Export Graph
Please refer to this section for more information.
2.4.4. Change Layout¶
Layouts provide a powerful means to organize nodes within a network. The following figures illustrate how a user may access and view automatically generated and previously saved layouts.
GraphSpace allows users to change layout using the following steps:
- Click on the
Change Layoutbutton to view available layout options. - The
Change Layoutpanel provides two alternatives: - Select Layout Algorithm - List of layout algorithms supported by GraphSpace through its use of Cytoscape.js. - Select Saved Layout - List of layout saved by the user using GraphSpace. The user has created them in earlier sessions by manually modifying the positions of nodes and edges created by some automatic layout algorithm and saving the layout. - Click on a layout option to change the current layout.
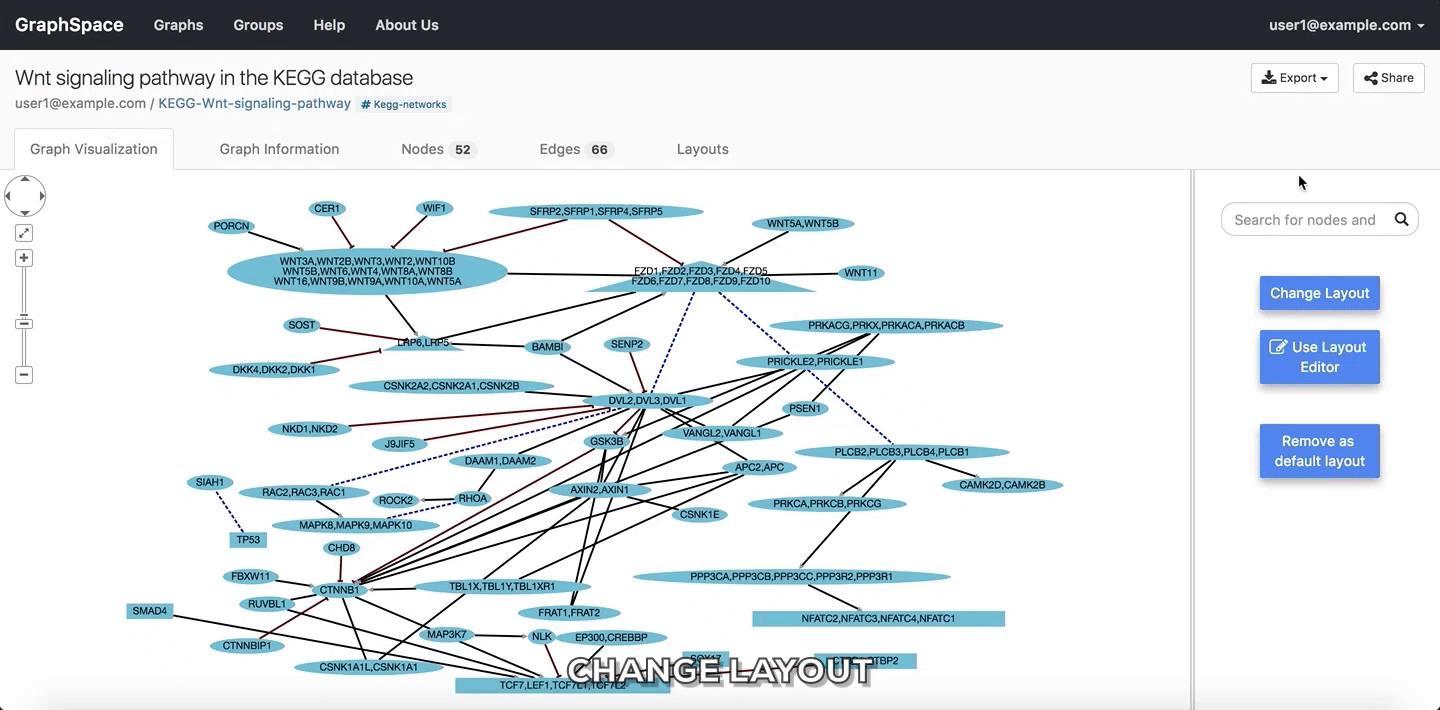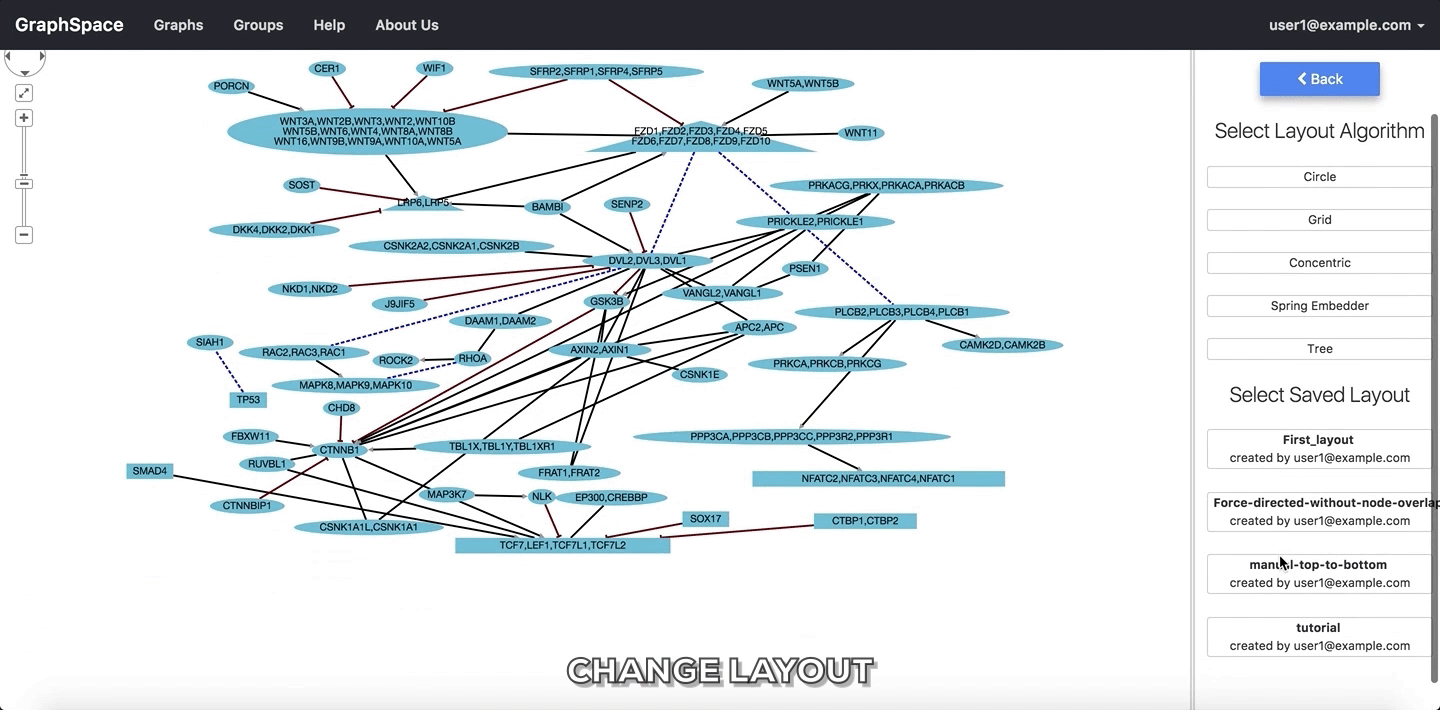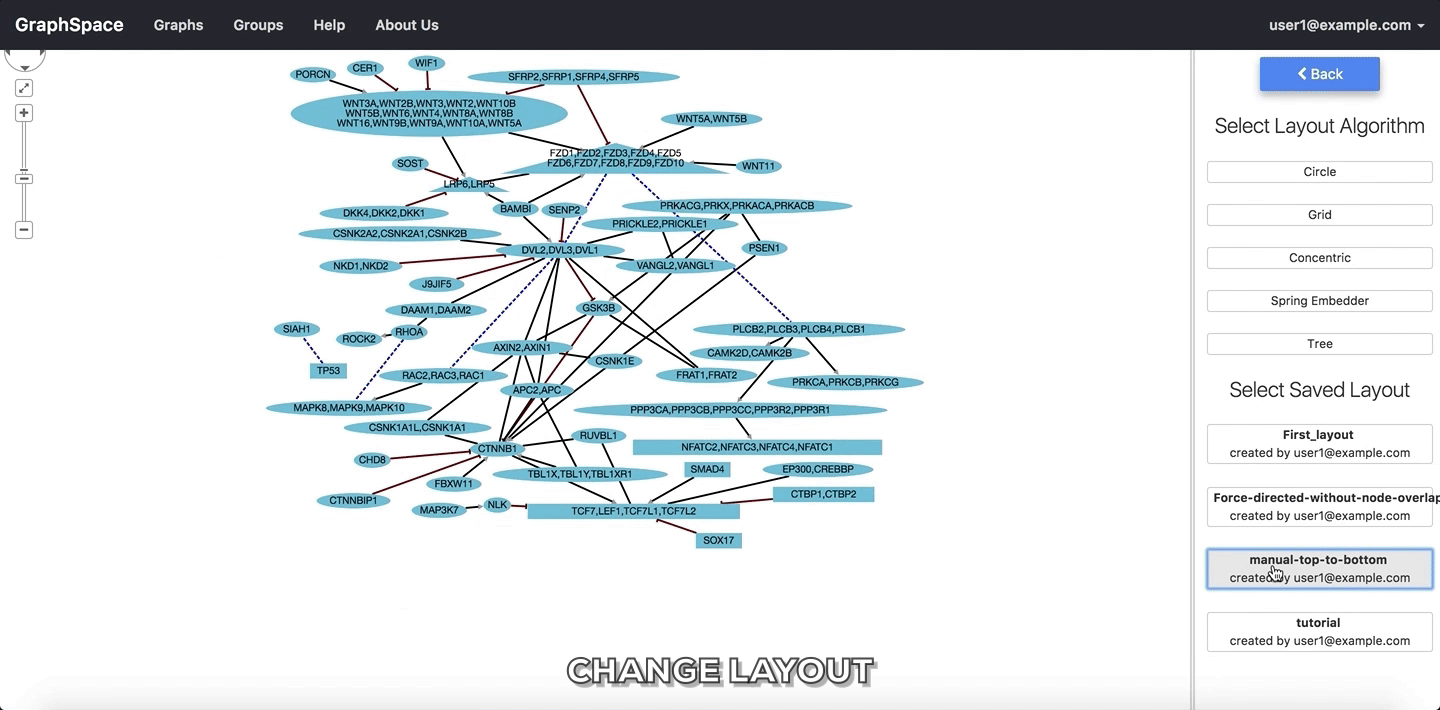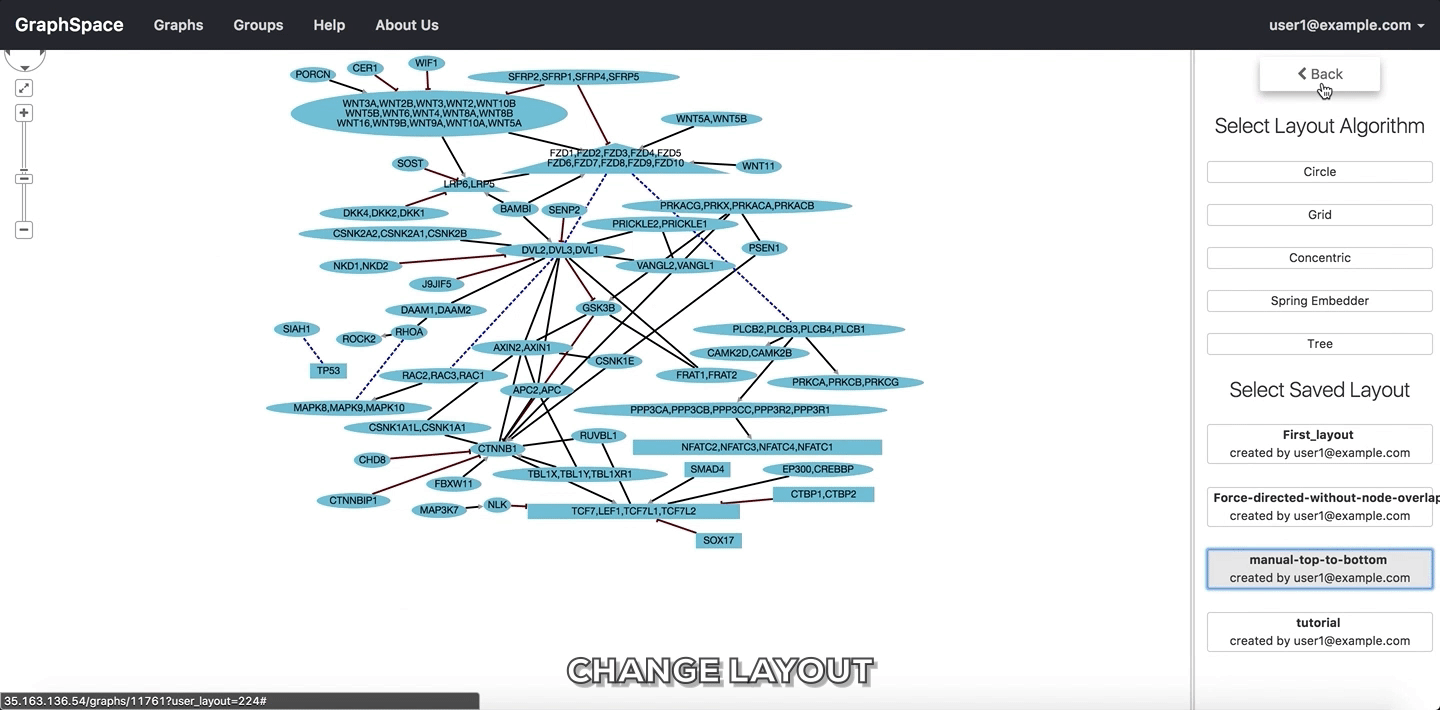
In the following example, the user selects to view the layout titled “manual-top-to-bottom”. To create this layout, the user manually moved nodes so that extracellular ligands and receptors (triangles) appear on the top while transcription factors (rectangles) appear at the bottom. The user can click on the Back button to return to the main menu.
 Change Layout
Change Layout
Please refer to this section for more information.
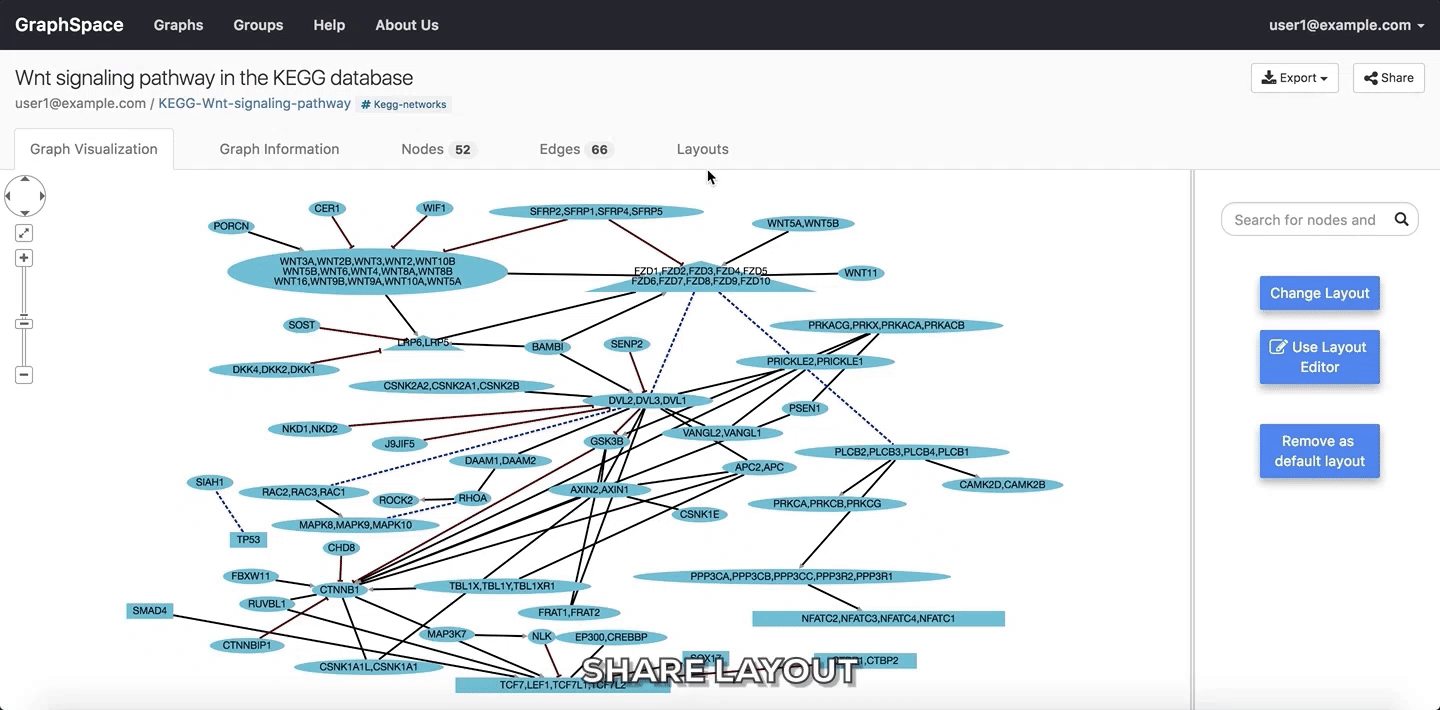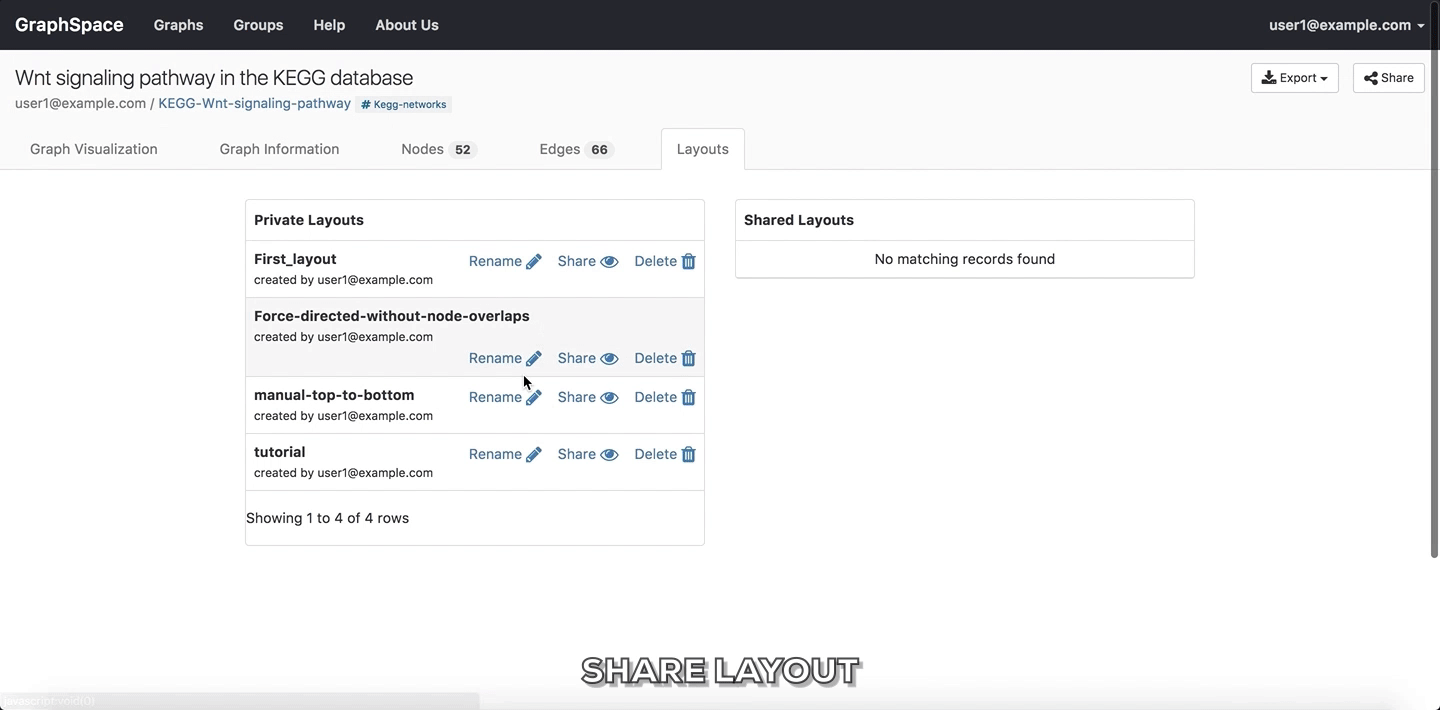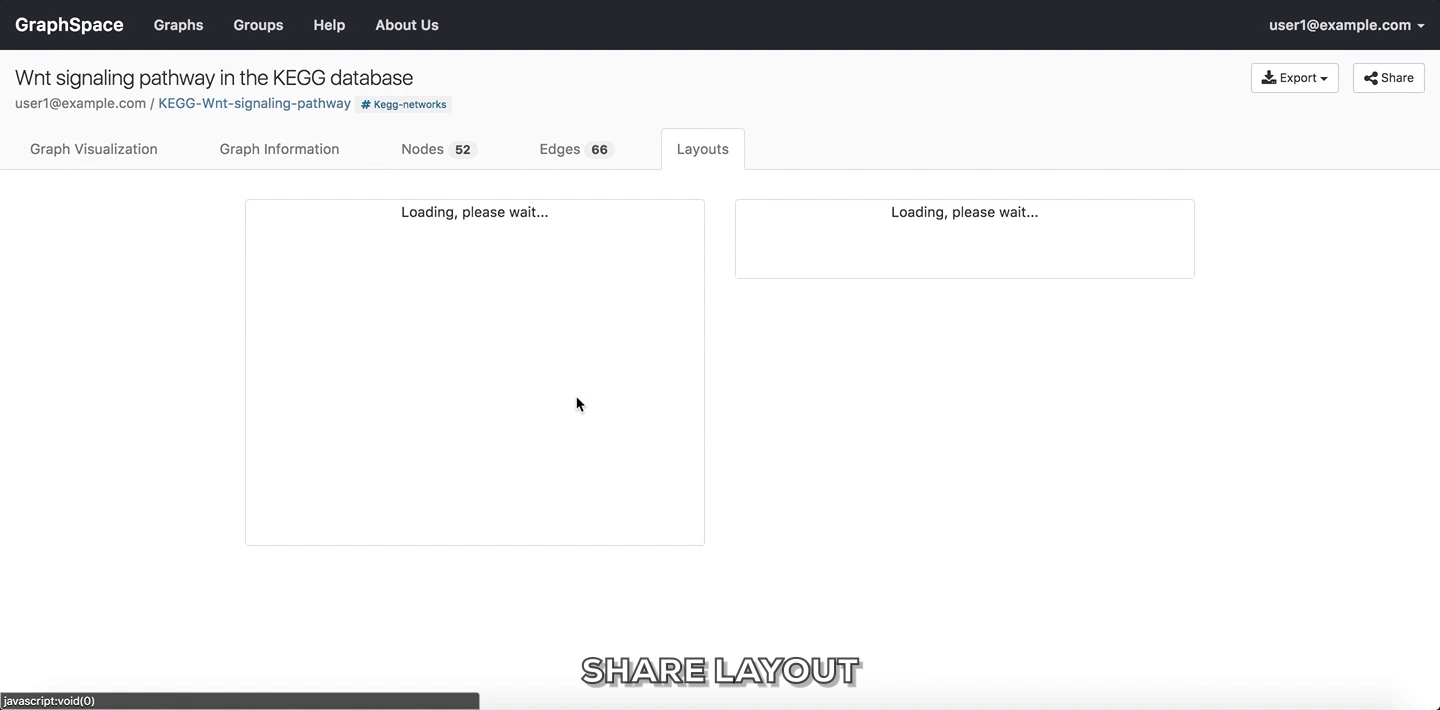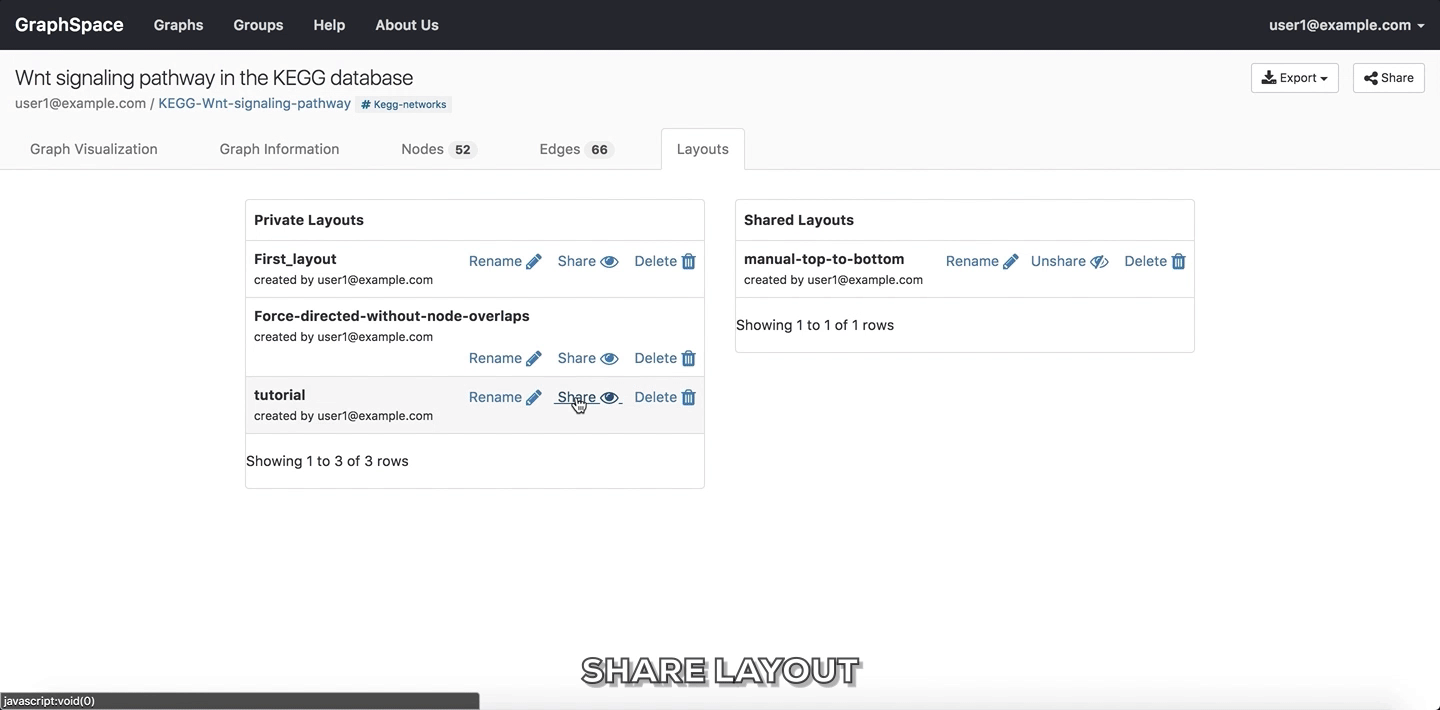 Share Layout
Share Layout